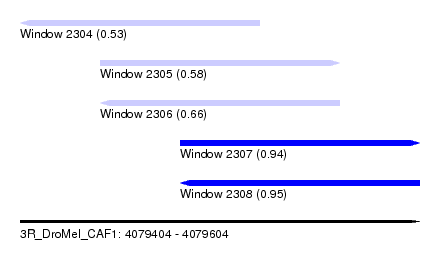
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,079,404 – 4,079,604 |
| Length | 200 |
| Max. P | 0.951551 |

| Location | 4,079,404 – 4,079,524 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.43 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4079404 120 - 27905053 AACUGCAACGGCUAAUUGCCGUUGAAAGUGUGUACAUUUUGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACCUGUCCCACGGGGCGUAUACUCACUAACGUAUUGGCUAUUAGCUGUCA .......(((((((((.((((......(.((((((...........((((((..(......)..))))))....((((....)))))))))).)((....))..)))).))))))))).. ( -33.90) >DroEre_CAF1 7932 120 - 1 AACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUUGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACUUGUCACACGGGGCGUAUACUCACUAACGAAUUGGCUAUUAGCUGUCA .(((((((.((((((((..(((((..((((((..(...........((((((..(......)..)))))).((((.....)))))..))))))...))))))))))))).)).))))).. ( -27.20) >DroYak_CAF1 4988 120 - 1 AACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUAGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACUUGUCACACGGGGCGUAUACUCACUAACGUAUUGGCUAUUAGCUGUCA .(((((((.(((((((...(((((..((((((..(...........((((((..(......)..)))))).((((.....)))))..))))))...))))).))))))).)).))))).. ( -26.90) >consensus AACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUUGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACUUGUCACACGGGGCGUAUACUCACUAACGUAUUGGCUAUUAGCUGUCA .(((((((.(((((((...(((((..((((((..(...........((((((..(......)..)))))).((((.....)))))..))))))...))))).))))))).)).))))).. (-26.54 = -26.43 + -0.11)



| Location | 4,079,444 – 4,079,564 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -22.14 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4079444 120 + 27905053 UGGGACAGGUAAACAUAAAAUAUUAUAAAUGUUUUCUGGCAAAAUGUACACACUUUCAACGGCAAUUAGCCGUUGCAGUUAAAUGUUUUUCUUAGCCAAACCUUUGGGAAUGGCUAAAAU .....((((.((((((............))))))))))(.(((((((....(((..(((((((.....))))))).)))...))))))).)(((((((..((....))..)))))))... ( -29.60) >DroEre_CAF1 7972 120 + 1 UGUGACAAGUAAACAUAAAAUAUUAUAAAUGUUUUCUGGCAAAAUGUACAUACUUUCAACGGCAAUUAGCCUUUGCCGUUGAAUGUUUUUCUUAGCCAAACCUUUGGGCCUCGGAAAGAU (((((((......((.(((((((.....))))))).))......))).))))..(((((((((((.......))))))))))).((((((((..(((.........)))...)))))))) ( -30.50) >DroYak_CAF1 5028 120 + 1 UGUGACAAGUAAACAUAAAAUAUUAUAAAUGUUUUCUGGCUAAAUGUACAUACUUUCAACGGCAAUUAGCCUUUGCCGUUGAAUGUUUUUCUUAGCCAAACCAUUGGGCCUCGGAAAGAU (((((((..((..((.(((((((.....))))))).))..))..))).))))..(((((((((((.......))))))))))).((((((((..(((.........)))...)))))))) ( -32.60) >consensus UGUGACAAGUAAACAUAAAAUAUUAUAAAUGUUUUCUGGCAAAAUGUACAUACUUUCAACGGCAAUUAGCCUUUGCCGUUGAAUGUUUUUCUUAGCCAAACCUUUGGGCCUCGGAAAGAU (((((((......((.(((((((.....))))))).))......))).))))..(((((((((((.......))))))))))).((((((((.((((((....))))..)).)))))))) (-22.14 = -23.70 + 1.56)



| Location | 4,079,444 – 4,079,564 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -22.27 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4079444 120 - 27905053 AUUUUAGCCAUUCCCAAAGGUUUGGCUAAGAAAAACAUUUAACUGCAACGGCUAAUUGCCGUUGAAAGUGUGUACAUUUUGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACCUGUCCCA .(((((((((..((....))..)))))))))...........((((((((((.....)))))))((((((....))))))..))).((((((..(......)..)))))).......... ( -33.30) >DroEre_CAF1 7972 120 - 1 AUCUUUCCGAGGCCCAAAGGUUUGGCUAAGAAAAACAUUCAACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUUGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACUUGUCACA .......((((.........((((((.((((...((((((((((((((.......))))))))))....))))...))))))))))((((((..(......)..)))))).))))..... ( -29.00) >DroYak_CAF1 5028 120 - 1 AUCUUUCCGAGGCCCAAUGGUUUGGCUAAGAAAAACAUUCAACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUAGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACUUGUCACA .......((((....((((.((((((((((....((((((((((((((.......))))))))))....))))...))))))))))...))))(((((....)))))....))))..... ( -32.20) >consensus AUCUUUCCGAGGCCCAAAGGUUUGGCUAAGAAAAACAUUCAACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUUGCCAGAAAACAUUUAUAAUAUUUUAUGUUUACUUGUCACA ....................((((((.((((...((((((((((((((.......))))))))))....))))...))))))))))((((((..(......)..)))))).......... (-22.27 = -22.50 + 0.23)


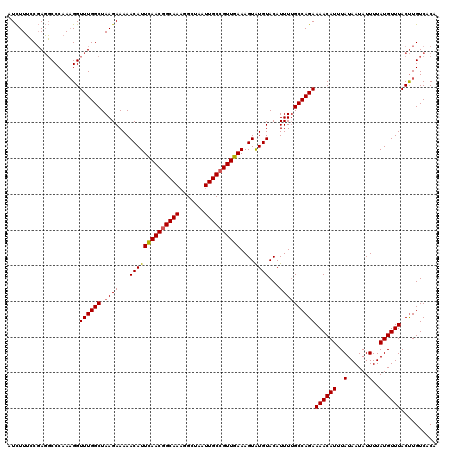
| Location | 4,079,484 – 4,079,604 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -29.40 |
| Energy contribution | -30.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4079484 120 + 27905053 AAAAUGUACACACUUUCAACGGCAAUUAGCCGUUGCAGUUAAAUGUUUUUCUUAGCCAAACCUUUGGGAAUGGCUAAAAUGCAUUGUUGCAAAUGCCACAGGGGCUAAAAGUACUAAAAA .....((((..(((..(((((((.....))))))).)))............((((((...((....))..((((.....((((....))))...))))....))))))..))))...... ( -34.10) >DroEre_CAF1 8012 120 + 1 AAAAUGUACAUACUUUCAACGGCAAUUAGCCUUUGCCGUUGAAUGUUUUUCUUAGCCAAACCUUUGGGCCUCGGAAAGAUGCAUUGUUGCAACUGCAAAAGUGGCUAAAAGUACUAAAAA .....((((..((.(((((((((((.......))))))))))).)).....(((((((.........((.((.....)).))....((((....))))...)))))))..))))...... ( -36.40) >DroYak_CAF1 5068 120 + 1 UAAAUGUACAUACUUUCAACGGCAAUUAGCCUUUGCCGUUGAAUGUUUUUCUUAGCCAAACCAUUGGGCCUCGGAAAGAUGCAUUGUUGCAACUGCAAAAGGGGCUAAAAGUACUAAAAA .....((((..((.(((((((((((.......))))))))))).)).....((((((...((....))(((.....((.((((....)))).)).....)))))))))..))))...... ( -37.60) >consensus AAAAUGUACAUACUUUCAACGGCAAUUAGCCUUUGCCGUUGAAUGUUUUUCUUAGCCAAACCUUUGGGCCUCGGAAAGAUGCAUUGUUGCAACUGCAAAAGGGGCUAAAAGUACUAAAAA .....((((..((.(((((((((((.......))))))))))).)).....((((((...((....))...........((((....))))...........))))))..))))...... (-29.40 = -30.07 + 0.67)

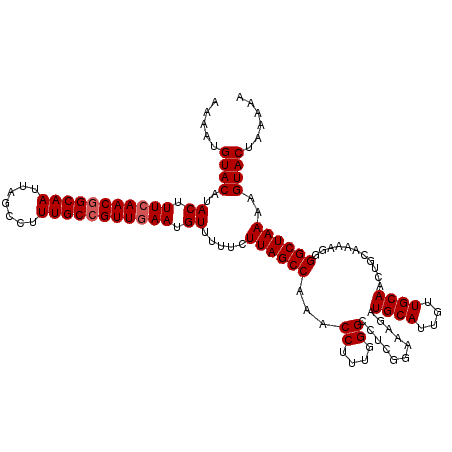

| Location | 4,079,484 – 4,079,604 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -32.01 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4079484 120 - 27905053 UUUUUAGUACUUUUAGCCCCUGUGGCAUUUGCAACAAUGCAUUUUAGCCAUUCCCAAAGGUUUGGCUAAGAAAAACAUUUAACUGCAACGGCUAAUUGCCGUUGAAAGUGUGUACAUUUU ......((((.....(((.....)))...((((....))))(((((((((..((....))..)))))))))...((((((.....(((((((.....))))))).))))))))))..... ( -38.90) >DroEre_CAF1 8012 120 - 1 UUUUUAGUACUUUUAGCCACUUUUGCAGUUGCAACAAUGCAUCUUUCCGAGGCCCAAAGGUUUGGCUAAGAAAAACAUUCAACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUU ......((((((((((((((((....((.((((....)))).))....)))(((....))).)))))))))...((.(((((((((((.......))))))))))).))..))))..... ( -37.90) >DroYak_CAF1 5068 120 - 1 UUUUUAGUACUUUUAGCCCCUUUUGCAGUUGCAACAAUGCAUCUUUCCGAGGCCCAAUGGUUUGGCUAAGAAAAACAUUCAACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUA ......((((((((((((((((....((.((((....)))).))....))))((....))...))))))))...((.(((((((((((.......))))))))))).))..))))..... ( -36.70) >consensus UUUUUAGUACUUUUAGCCCCUUUUGCAGUUGCAACAAUGCAUCUUUCCGAGGCCCAAAGGUUUGGCUAAGAAAAACAUUCAACGGCAAAGGCUAAUUGCCGUUGAAAGUAUGUACAUUUU ......((((((((((((...((((....((((....))))......))))(((....)))..))))))))...((.(((((((((((.......))))))))))).))..))))..... (-32.01 = -31.57 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:09 2006