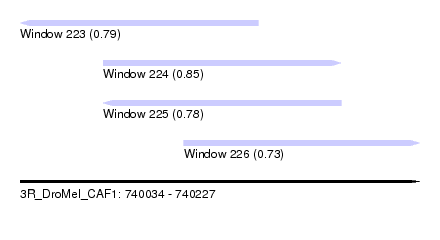
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 740,034 – 740,227 |
| Length | 193 |
| Max. P | 0.848553 |

| Location | 740,034 – 740,149 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.82 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -25.60 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
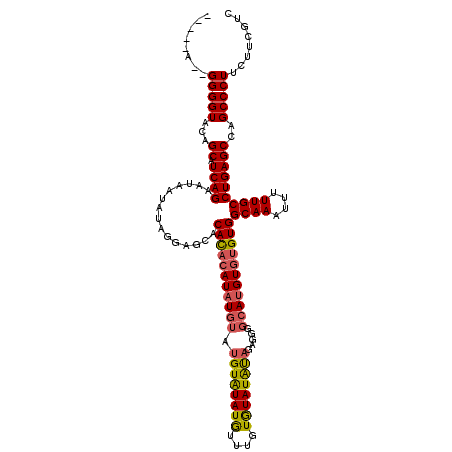
>3R_DroMel_CAF1 740034 115 - 27905053 UACAUAU----GUAUAUGUUUGUAUAUGCAGAGGGACAUGUGUGUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUCUGCAUUUCUACUAUUCAGCAAGUGAGCAUAUGAUGUUCCU ......(----(((((((....)))))))).(((((((((((((((((((....)))))...............(((..(((..............)))..))))))))).)))))))) ( -30.34) >DroSec_CAF1 7406 111 - 1 CACAUUUGUAUGUGUAUAUUUGUGUAUAUAGAGGGGCAUGUGUGUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUCUGCAUUUCUACUAGUCA--------GCAUAUGAUGUUCCU .(((.(..((((((((.(..(((..((..((((((((.((.((..(((((....)))))...))))))))))))..))..)))..).)))......--------)))))..)))).... ( -32.30) >DroSim_CAF1 7415 109 - 1 CACAUAUGUAUGUAUAUGUUUGUGUAUAUAGAGGGGCAUGU--GUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUCUGCAUUUCUACUAGUCA--------GCAUAUGAUGUUCCU ..(((((((((((((((....))))))))((((((((..((--..(((((....)))))...))..))))))))......................--------)))))))........ ( -32.60) >consensus CACAUAUGUAUGUAUAUGUUUGUGUAUAUAGAGGGGCAUGUGUGUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUCUGCAUUUCUACUAGUCA________GCAUAUGAUGUUCCU ..(((((((..(((((((....)))))))((((((((..((....(((((....)))))...))..))))))))..............................)))))))........ (-25.60 = -25.83 + 0.23)

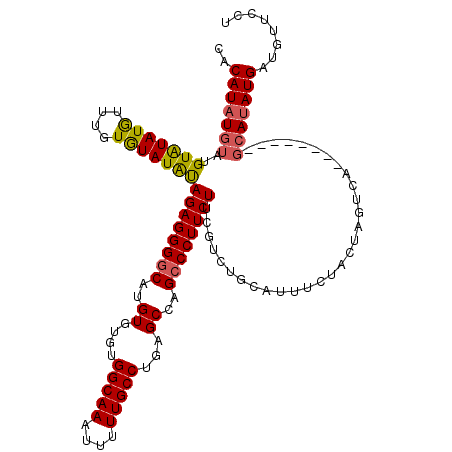
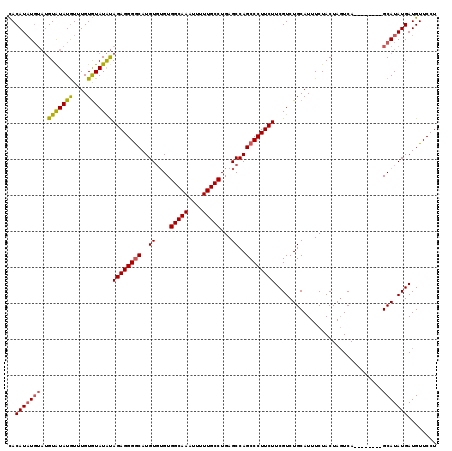
| Location | 740,074 – 740,189 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 740074 115 + 27905053 GACGAAGAAGGGCUGGCUCAGGCAAAAAUUUGCCACACACAUGUCCCUCUGCAUAUACAAACAUAUAC----AUAUGUAUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCCCCUAUACC .....((..(((..(((((((((((....)))))................(((((((((.........----...))))))))).............))).)))...)))..))..... ( -25.00) >DroSec_CAF1 7438 112 + 1 GACGAAGAAGGGCUGGCUCAGGCAAAAAUUUGCCACACACAUGCCCCUCUAUAUACACAAAUAUACACAUACAAAUGUGUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCC--U----- .....(((.((((((.....(((((....))))).....)).)))).)))...(((((((((((((((((((....)))))))....)))))).......)).))))....--.----- ( -26.91) >DroSim_CAF1 7447 110 + 1 GACGAAGAAGGGCUGGCUCAGGCAAAAAUUUGCCAC--ACAUGCCCCUCUAUAUACACAAACAUAUACAUACAUAUGUGUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCC--U----- .....(((.((((((.....(((((....)))))..--.)).)))).))).....((((.((((((......)))))).))))............................--.----- ( -24.80) >consensus GACGAAGAAGGGCUGGCUCAGGCAAAAAUUUGCCACACACAUGCCCCUCUAUAUACACAAACAUAUACAUACAUAUGUGUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCC__U_____ .....(((.((((((.....(((((....))))).....)).)))).)))...........(((((((((....))))))))).................................... (-23.55 = -23.33 + -0.22)

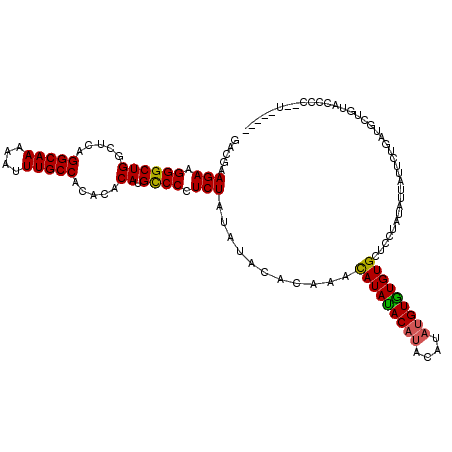
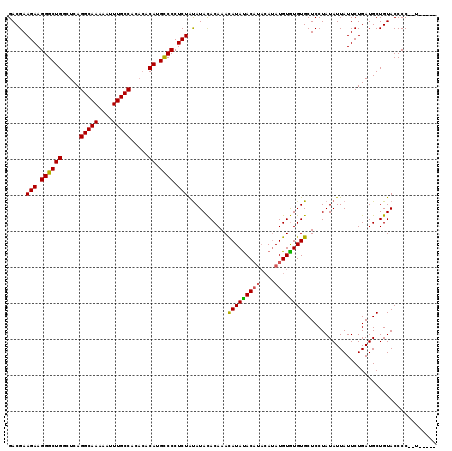
| Location | 740,074 – 740,189 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -28.55 |
| Energy contribution | -29.23 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 740074 115 - 27905053 GGUAUAGGGGGGUACAGCAUCAGAAUAAUAUAGGAGCACAUACAUAU----GUAUAUGUUUGUAUAUGCAGAGGGACAUGUGUGUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUC .....((((((((...((.((((...............(((((((((----((...(.(((((....))))).).)))))))))))((((....))))))))))..))))))))..... ( -37.40) >DroSec_CAF1 7438 112 - 1 -----A--GGGGUACAGCAUCAGAAUAAUAUAGGAGCACACACAUUUGUAUGUGUAUAUUUGUGUAUAUAGAGGGGCAUGUGUGUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUC -----(--(((((...((.((((.............((((((((..(((.(.(((((((....))))))).)...)))))))))))((((....))))))))))..))))))....... ( -33.90) >DroSim_CAF1 7447 110 - 1 -----A--GGGGUACAGCAUCAGAAUAAUAUAGGAGCACACACAUAUGUAUGUAUAUGUUUGUGUAUAUAGAGGGGCAUGU--GUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUC -----.--..............(((...((((...(((((.(((((((....))))))).))))).))))(((((((..((--..(((((....)))))...))..))))))))))... ( -33.20) >consensus _____A__GGGGUACAGCAUCAGAAUAAUAUAGGAGCACACACAUAUGUAUGUAUAUGUUUGUGUAUAUAGAGGGGCAUGUGUGUGGCAAAUUUUUGCCUGAGCCAGCCCUUCUUCGUC ........(((((...((.((((...............(((((((((((.((((((((....)))))))).....)))))))))))((((....))))))))))..)))))........ (-28.55 = -29.23 + 0.68)

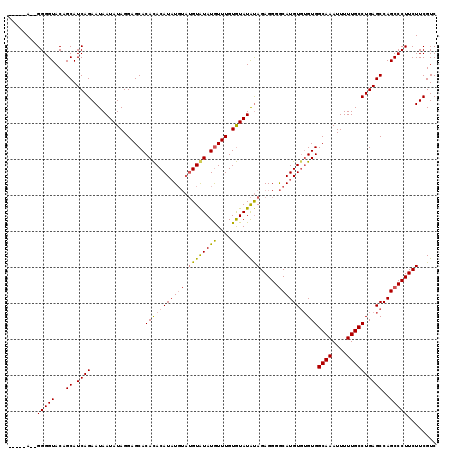
| Location | 740,113 – 740,227 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.16 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.17 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 740113 114 + 27905053 CAUGUCCCUCUGCAUAUACAAACAUAUAC----AUAUGUAUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCCCCUAUACCCUAUACAUACCACCAAGUGUGUGCCAUAAAGAAAGGCU ...(((..((((((((((((.........----...)))))))))......................................(((((((......)))))))......)))..))). ( -19.00) >DroSec_CAF1 7477 105 + 1 CAUGCCCCUCUAUAUACACAAAUAUACACAUACAAAUGUGUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCC--U-----------AUACCUCGAAGUGUGUGCCAUAAAGAAAGGCU ...(((..(((.(((((((.(((((((((((((....)))))))....)))))).(((.((.(.((((....--)-----------)))).))))))))))))......)))..))). ( -21.50) >DroSim_CAF1 7484 105 + 1 CAUGCCCCUCUAUAUACACAAACAUAUACAUACAUAUGUGUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCC--U-----------AUACCUCGAAGUGUGUGCCAUAAAGAAAGGCU ...(((..(((.(((((((...(((((((((....)))))))))...........(((.((.(.((((....--)-----------)))).))))))))))))......)))..))). ( -20.30) >consensus CAUGCCCCUCUAUAUACACAAACAUAUACAUACAUAUGUGUGUGCUCCUAUAUUAUUCUGAUGCUGUACCCC__U___________AUACCUCGAAGUGUGUGCCAUAAAGAAAGGCU ...(((..(((.(((((((...(((((((((....)))))))))....((((.(((....))).))))............................)))))))......)))..))). (-17.82 = -17.17 + -0.66)

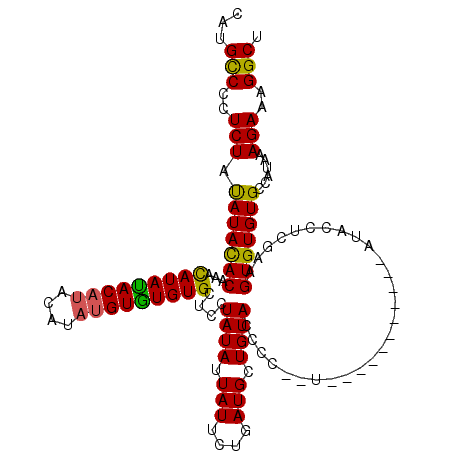
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:11 2006