
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,053,992 – 4,054,123 |
| Length | 131 |
| Max. P | 0.968645 |

| Location | 4,053,992 – 4,054,098 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -24.48 |
| Energy contribution | -25.14 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
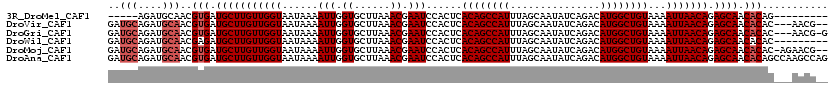
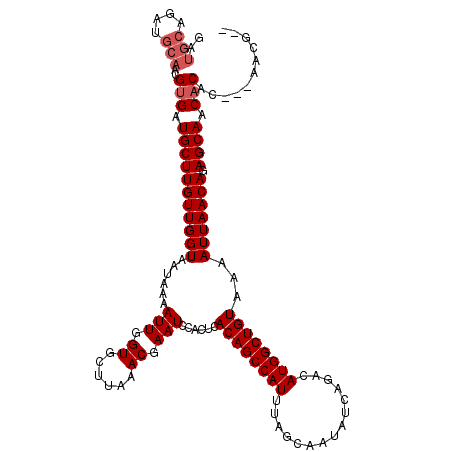
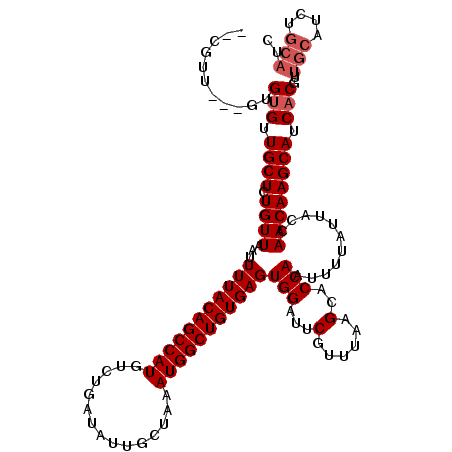
>3R_DroMel_CAF1 4053992 106 + 27905053 -----AGAUGCAACGUGAUGCUUGUUGGUAAUAAAAUUGGUGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAG--------- -----.........(((.(((((((((((......(((.((......)).)))......((((((((...............))))))))...))))))).)))).)))..--------- ( -24.96) >DroVir_CAF1 39861 115 + 1 GAUGCAGAUGCAACGUGAUGCUUGUUGGUAAUAAAAUUGGUGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC---AACG-- ..(((....)))..(((.(((((((((((......(((.((......)).)))......((((((((...............))))))))...))))))).)))).)))..---....-- ( -27.26) >DroGri_CAF1 23294 116 + 1 GAUGCAGAUGCAACGUGAUGCUUGUUGGUAAUAAAAUUGGUGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC---AACG-G ..(((....)))..(((.(((((((((((......(((.((......)).)))......((((((((...............))))))))...))))))).)))).)))..---....-. ( -27.26) >DroWil_CAF1 70476 111 + 1 GAUGCAGAUGCAACGAGAUGCUUGUUGGUAAUAAAAUUGGUGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC--------- ..(((....)))....(.(((((((((((......(((.((......)).)))......((((((((...............))))))))...))))))).)))).)....--------- ( -22.86) >DroMoj_CAF1 45874 117 + 1 GAUGCAGAUGCAACGUGAUGCUUGUUGGUAAUAAAAUUGGUGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC-AGAACG-- ..(((....)))..(((.(((((((((((......(((.((......)).)))......((((((((...............))))))))...))))))).)))).)))..-......-- ( -27.26) >DroAna_CAF1 44939 120 + 1 GAUGCAGAUGCAACGUGAUGCUUGUUGGUAAUAAAAUUGGUGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAGCCAAGCCAG ..(((....)))..(((.(((((((((((......(((.((......)).)))......((((((((...............))))))))...))))))).)))).)))........... ( -27.26) >consensus GAUGCAGAUGCAACGUGAUGCUUGUUGGUAAUAAAAUUGGUGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC___AACG__ ..(((....)))..(((.(((((((((((......(((.((......)).)))......((((((((...............))))))))...))))))).)))).)))........... (-24.48 = -25.14 + 0.67)
| Location | 4,053,992 – 4,054,098 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -24.43 |
| Energy contribution | -25.09 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
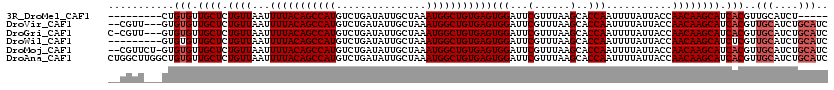
>3R_DroMel_CAF1 4053992 106 - 27905053 ---------CUGUGUUGCUCUGUUAAUUUUACAGCCAUGUCUGAUAUUGCUAAAUGGCUGUGAGUGGAUUCGUUUAAGCACCAAUUUUAUUACCAACAAGCAUCACGUUGCAUCU----- ---------..(((.((((.((((...(((((((((((...............)))))))))))(((...(......)..)))...........)))))))).))).........----- ( -25.16) >DroVir_CAF1 39861 115 - 1 --CGUU---GUGUGUUGCUCUGUUAAUUUUACAGCCAUGUCUGAUAUUGCUAAAUGGCUGUGAGUGGAUUCGUUUAAGCACCAAUUUUAUUACCAACAAGCAUCACGUUGCAUCUGCAUC --.((.---.((((.((((.((((...(((((((((((...............)))))))))))(((...(......)..)))...........)))))))).))))..))......... ( -28.06) >DroGri_CAF1 23294 116 - 1 C-CGUU---GUGUGUUGCUCUGUUAAUUUUACAGCCAUGUCUGAUAUUGCUAAAUGGCUGUGAGUGGAUUCGUUUAAGCACCAAUUUUAUUACCAACAAGCAUCACGUUGCAUCUGCAUC .-.((.---.((((.((((.((((...(((((((((((...............)))))))))))(((...(......)..)))...........)))))))).))))..))......... ( -28.06) >DroWil_CAF1 70476 111 - 1 ---------GUGUGUUGCUCUGUUAAUUUUACAGCCAUGUCUGAUAUUGCUAAAUGGCUGUGAGUGGAUUCGUUUAAGCACCAAUUUUAUUACCAACAAGCAUCUCGUUGCAUCUGCAUC ---------..(.(.((((.((((...(((((((((((...............)))))))))))(((...(......)..)))...........)))))))).).)..(((....))).. ( -23.16) >DroMoj_CAF1 45874 117 - 1 --CGUUCU-GUGUGUUGCUCUGUUAAUUUUACAGCCAUGUCUGAUAUUGCUAAAUGGCUGUGAGUGGAUUCGUUUAAGCACCAAUUUUAUUACCAACAAGCAUCACGUUGCAUCUGCAUC --......-..(((.((((.((((...(((((((((((...............)))))))))))(((...(......)..)))...........)))))))).)))..(((....))).. ( -27.16) >DroAna_CAF1 44939 120 - 1 CUGGCUUGGCUGUGUUGCUCUGUUAAUUUUACAGCCAUGUCUGAUAUUGCUAAAUGGCUGUGAGUGGAUUCGUUUAAGCACCAAUUUUAUUACCAACAAGCAUCACGUUGCAUCUGCAUC ...((...((((((.((((.((((...(((((((((((...............)))))))))))(((...(......)..)))...........)))))))).))))..))....))... ( -29.16) >consensus __CGUU___GUGUGUUGCUCUGUUAAUUUUACAGCCAUGUCUGAUAUUGCUAAAUGGCUGUGAGUGGAUUCGUUUAAGCACCAAUUUUAUUACCAACAAGCAUCACGUUGCAUCUGCAUC ...........(((.((((.((((...(((((((((((...............)))))))))))(((...(......)..)))...........)))))))).)))..(((....))).. (-24.43 = -25.09 + 0.67)
| Location | 4,054,027 – 4,054,123 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -18.89 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.56 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
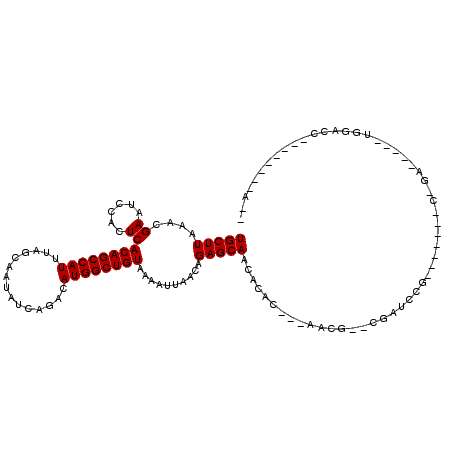
>3R_DroMel_CAF1 4054027 96 + 27905053 UGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAG---------CGAUCCG-------C-GA-----UGGAGCAACUGGCAA-- ((((........((((.((((((((((...............))))))))....................(---------((...))-------)-))-----))))......)))).-- ( -21.30) >DroVir_CAF1 39901 92 + 1 UGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC---AACG--------------AAC-GA-----UGCUCCAA---ACAA-- ...................((((((((...............))))))))..........(((((.(....---....--------------...-).-----)))))...---....-- ( -14.76) >DroGri_CAF1 23334 99 + 1 UGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC---AACG-GCGACCCGGCCGAAAC-GA-----UGGACC----------- ............((((.((((((((((...............)))))))).....................---..((-((......))))....-))-----))))..----------- ( -22.06) >DroMoj_CAF1 45914 92 + 1 UGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC-AGAACG--------------AGC-GA-----UGGACCCA---A--A-- .........(..((((.((((((((((...............))))))))............((.......-......--------------.))-))-----))))..).---.--.-- ( -15.20) >DroAna_CAF1 44979 93 + 1 UGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAGCCAAGCCAGCGAUCCA-------G-CA-------------------ACC (((((....((......))((((((((...............))))))))..........)))))..............((......-------)-).-------------------... ( -16.66) >DroPer_CAF1 40425 100 + 1 UGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAG---------CGAUGCG-------GCCAGAGCGUGGCGC----GGCAAUC (((((....((......))((((((((...............))))))))..........))))).....(---------(.((((.-------......)))).))..----....... ( -23.36) >consensus UGCUUAAACGAAUCCACUCACAGCCAUUUAGCAAUAUCAGACAUGGCUGUAAAAUUAACAGAGCAACACAC___AACG__CGAUCCG_______C_GA_____UGGACC________A__ (((((....((......))((((((((...............))))))))..........)))))....................................................... (-14.56 = -14.56 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:50 2006