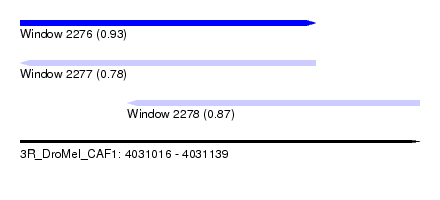
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,031,016 – 4,031,139 |
| Length | 123 |
| Max. P | 0.925936 |

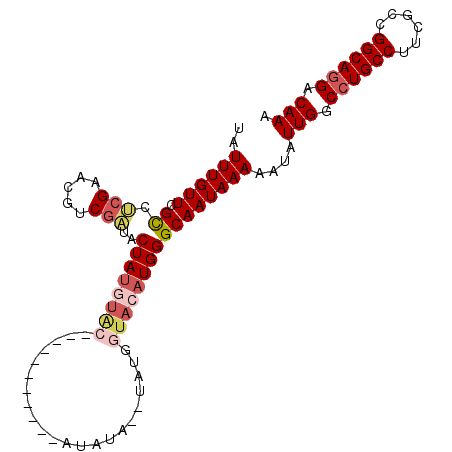
| Location | 4,031,016 – 4,031,107 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.90 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4031016 91 + 27905053 UAUUUGUUCGCCUCGAACGUCGAUACUAUGUAC-----------AUAUAUAUAUACUACAUGGGCAAUAAAAAUAUUGGCCUGCCUUCGCCGGCAGGACAAA ..((((((.((((((.....)))...((((((.-----------.(((....))).)))))))))))))))....(((.((((((......)))))).))). ( -23.90) >DroSec_CAF1 36534 89 + 1 UAUUUGUUCGCCUCGAACAUCGAUACUAUGUAC-----------AUAUA--UUUGGUACAUGGGCAAUAAAAAUAUUGGCCUGCCUUCGCCGGCAUGACAAA ..((((((.((((((.....)))..((((((((-----------.....--....))))))))..............))).((((......)))).)))))) ( -22.10) >DroSim_CAF1 37075 89 + 1 UAUUUGUUCGCCUCGAACAUCGAUACUAUGUAC-----------AUAUA--UAUGGUACAUGGGCAAUAAAAAUAUUGGCCUGCCUUCGCCGGCAGGACAAA ..((((((.((((((.....)))(((((((((.-----------...))--)))))))....)))))))))....(((.((((((......)))))).))). ( -28.40) >DroEre_CAF1 19653 100 + 1 UAUUUGUUCGCCCCGAACGCCGAUACUAUGUGCGGGUAUAUGGCGUAUA--UAUGGUACAUGGGCAAUAAAAAUAUUGGCCUGCCUUCGCCGGCAGGACAAA ..((((((.((((.....(((((((((.......))))).))))((((.--....))))..))))))))))....(((.((((((......)))))).))). ( -34.50) >DroYak_CAF1 37579 93 + 1 UAUUUGUUCGCCUCGAACGUCGAUACUAUAGGC---UGUAUGCUG------UAUGGUACAUGGGCAAUAAAAAUAUUGGCCUGCCUUCGCCGGCAGGACAAA ..((((((.((((((.....)))((((((((((---.....))).------)))))))....)))))))))....(((.((((((......)))))).))). ( -30.20) >DroAna_CAF1 20313 82 + 1 UAUUUGUUAGUAGUGG----CCCUGCUACAUGC----------UA------CAUACAACAUGGGCAAUAAAAAUAUUGGCCUGCCUUCGCCGGCAGGACAAA ..((((((.(((((..----....)))))..((----------(.------(((.....))))))))))))....(((.((((((......)))))).))). ( -23.10) >consensus UAUUUGUUCGCCUCGAACGUCGAUACUAUGUAC___________AUAUA__UAUGGUACAUGGGCAAUAAAAAUAUUGGCCUGCCUUCGCCGGCAGGACAAA ..((((((.((.(((.....)))..((((((((......................))))))))))))))))....(((.((((((......)))))).))). (-18.76 = -19.90 + 1.14)


| Location | 4,031,016 – 4,031,107 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.79 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4031016 91 - 27905053 UUUGUCCUGCCGGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAUGUAGUAUAUAUAUAU-----------GUACAUAGUAUCGACGUUCGAGGCGAACAAAUA .(((.((((((......)))))).)))........(((((......((((((....))-----------))))......(((.....))))))))....... ( -26.00) >DroSec_CAF1 36534 89 - 1 UUUGUCAUGCCGGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAUGUACCAAA--UAUAU-----------GUACAUAGUAUCGAUGUUCGAGGCGAACAAAUA (((((...(((.((....)).)))...........(((((.((((((....--.....-----------))))))....(((.....))))))))))))).. ( -22.60) >DroSim_CAF1 37075 89 - 1 UUUGUCCUGCCGGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAUGUACCAUA--UAUAU-----------GUACAUAGUAUCGAUGUUCGAGGCGAACAAAUA .(((.((((((......)))))).)))........(((((.((((((....--.....-----------))))))....(((.....))))))))....... ( -26.00) >DroEre_CAF1 19653 100 - 1 UUUGUCCUGCCGGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAUGUACCAUA--UAUACGCCAUAUACCCGCACAUAGUAUCGGCGUUCGGGGCGAACAAAUA .(((.((((((......)))))).)))........(((((((((((...))--)))(((((..((((.........)))).)))))...))))))....... ( -30.70) >DroYak_CAF1 37579 93 - 1 UUUGUCCUGCCGGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAUGUACCAUA------CAGCAUACA---GCCUAUAGUAUCGACGUUCGAGGCGAACAAAUA (((((..((((.((..((((.((.(((((....))))).))((((......------..))))...---))))...)).(((.....))))))).))))).. ( -24.40) >DroAna_CAF1 20313 82 - 1 UUUGUCCUGCCGGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAUGUUGUAUG------UA----------GCAUGUAGCAGGG----CCACUACUAACAAAUA .(((.((((((......)))))).)))..........((((.(((((((((------..----------.))))))))))))----)............... ( -26.40) >consensus UUUGUCCUGCCGGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAUGUACCAUA__UAUAU___________GCACAUAGUAUCGACGUUCGAGGCGAACAAAUA .(((.((((((......)))))).)))........(((((.((((..........................))))....(((.....))))))))....... (-15.53 = -15.79 + 0.25)

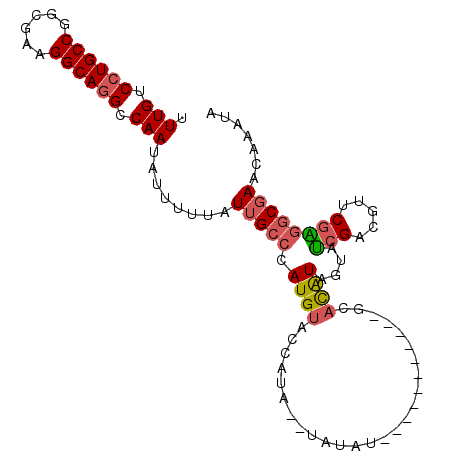
| Location | 4,031,049 – 4,031,139 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -13.88 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4031049 90 - 27905053 -GAAAUUGUGCAUGC--AUUUUAUGGUGCAAUUCCUUUGUCCUGCC------GGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAU--GUAGUA-----UAUAUAUAU -.....((((.((((--....(((((.(((((....(((.((((((------......)))))).))).......)))))))))--)..)))-----).))))... ( -27.60) >DroPse_CAF1 17929 100 - 1 -CAUAUUGUGUAUAC--AUUUUAUGGUGCAAUUCCUUUGUCCUCGC--AG-CGCCGAAGGCAGGCCAAUAUUUUUAUUGCUCGCUUCUGCCAGAGCAACUAUCCAU -((((..(((....)--))..))))...........(((..((.((--((-......((((..((.((((....))))))..)))))))).))..)))........ ( -21.40) >DroSec_CAF1 36567 88 - 1 -GAAAUUGUGCAUGC--AUUUUAUGGUGCAAUUCCUUUGUCAUGCC------GGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAU--GUACCA-----AA--UAUAU -.(((.(((....))--).))).(((((((...((((((((.....------))))))))..((.(((((....))))).)).)--))))))-----..--..... ( -25.80) >DroSim_CAF1 37108 88 - 1 -GAAAUUGUGCAUGC--AUUUUAUGGUGCAAUUCCUUUGUCCUGCC------GGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAU--GUACCA-----UA--UAUAU -......(((....)--))..(((((((((...((((((((.....------))))))))..((.(((((....))))).)).)--))))))-----))--..... ( -26.50) >DroWil_CAF1 46295 98 - 1 UCAGAUUGUGUAUGCACAUUUUAUGGUGCAAUUCUUUUGUGCUCUCGCACUUGGCGAAGGCAGGCCAAUAUUUUUAUUACCCGCUUGUACAAGA----A--UUU-- ............(((((........)))))....((((((((..((((.....))))((((.((..((((....)))).)).))))))))))))----.--...-- ( -24.00) >DroPer_CAF1 18235 100 - 1 -CAUAUUGUGUAUAC--AUUUUAUGGUGCAAUUCCUUUGUCCUCGC--AG-CGCCGAAGGCAGGCCAAUAUUUUUAUUGCUCGCUUCUGCCAGAGCAACUAUCCAU -((((..(((....)--))..))))...........(((..((.((--((-......((((..((.((((....))))))..)))))))).))..)))........ ( -21.40) >consensus _CAAAUUGUGCAUGC__AUUUUAUGGUGCAAUUCCUUUGUCCUCCC______GGCGAAGGCAGGCCAAUAUUUUUAUUGCCCAC__GUACCA_____AA__UAUAU .......................((((((....((((((((...........))))))))..((.(((((....))))).))....)))))).............. (-13.88 = -14.52 + 0.64)

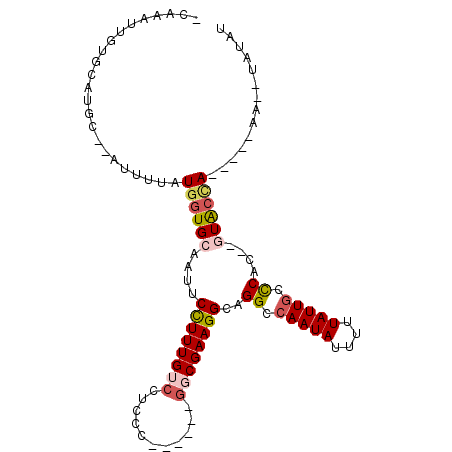
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:39 2006