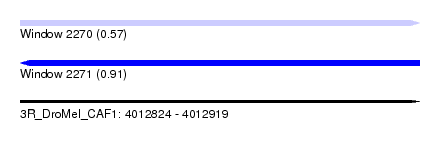
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,012,824 – 4,012,919 |
| Length | 95 |
| Max. P | 0.909679 |

| Location | 4,012,824 – 4,012,919 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -28.03 |
| Energy contribution | -28.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4012824 95 + 27905053 UUGGAGCUAGUGCUCCUG------------------------UUCCUGCUAAAGAAAGCUUGGCUAAGCACCUGUGCAAACUUAUUGUUAGCCUGCGCAUUGUGUUCGAUCGAAGCGGA ..(((((....)))))..------------------------...(((((.......((..((((..(((....))).(((.....))))))).))(.((((....)))))..))))). ( -27.40) >DroSec_CAF1 18506 95 + 1 UUGGAGCUAGGGCUCCUG------------------------CUCCUGCUAACGAAAGCUUGGCUAAGCACCUGUGCAAACUUAUUGUUAGCCUGCGCAUUGUGUUCGAUCCAAGCGGA ..(((((.(((...))))------------------------))))...........((((((...(((((.((((((..((.......))..))))))..)))))....))))))... ( -31.90) >DroSim_CAF1 19088 119 + 1 UUGGAGCUAGGGCUCCUGCUCCUGCUAGUGCUACUUCUCCUGCUCCUGCUAACGAAAGCUUGGCUAAGCACCUGUGCAAACUUAUUGUUAGCCUGCGCAUUGUGUUCGAUCCAAGCGGA ..(((((.(((((....)).)))((....))..........)))))...........((((((...(((((.((((((..((.......))..))))))..)))))....))))))... ( -36.60) >consensus UUGGAGCUAGGGCUCCUG________________________CUCCUGCUAACGAAAGCUUGGCUAAGCACCUGUGCAAACUUAUUGUUAGCCUGCGCAUUGUGUUCGAUCCAAGCGGA ..(((((....))))).........................................((((((...(((((.((((((..((.......))..))))))..)))))....))))))... (-28.03 = -28.37 + 0.33)


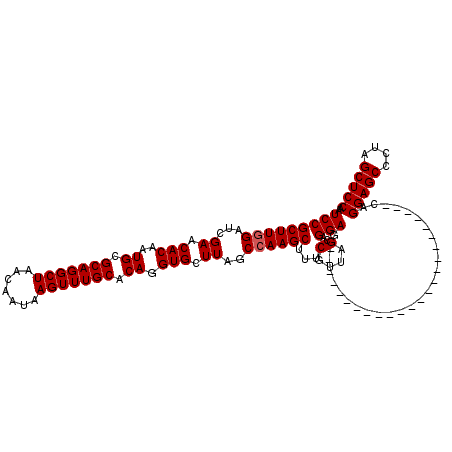
| Location | 4,012,824 – 4,012,919 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -28.47 |
| Energy contribution | -28.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4012824 95 - 27905053 UCCGCUUCGAUCGAACACAAUGCGCAGGCUAACAAUAAGUUUGCACAGGUGCUUAGCCAAGCUUUCUUUAGCAGGAA------------------------CAGGAGCACUAGCUCCAA ((((((((....)).(((..((.(((((((.......))))))).)).)))...)))...(((......))).))).------------------------..(((((....))))).. ( -27.20) >DroSec_CAF1 18506 95 - 1 UCCGCUUGGAUCGAACACAAUGCGCAGGCUAACAAUAAGUUUGCACAGGUGCUUAGCCAAGCUUUCGUUAGCAGGAG------------------------CAGGAGCCCUAGCUCCAA ...((((((...((.(((..((.(((((((.......))))))).)).))).))..))))))...........((((------------------------((((...))).))))).. ( -32.80) >DroSim_CAF1 19088 119 - 1 UCCGCUUGGAUCGAACACAAUGCGCAGGCUAACAAUAAGUUUGCACAGGUGCUUAGCCAAGCUUUCGUUAGCAGGAGCAGGAGAAGUAGCACUAGCAGGAGCAGGAGCCCUAGCUCCAA (((((((((...((.(((..((.(((((((.......))))))).)).))).))..))))))........((....)).)))......((....)).((((((((...))).))))).. ( -38.50) >consensus UCCGCUUGGAUCGAACACAAUGCGCAGGCUAACAAUAAGUUUGCACAGGUGCUUAGCCAAGCUUUCGUUAGCAGGAG________________________CAGGAGCCCUAGCUCCAA (((((((((...((.(((..((.(((((((.......))))))).)).))).))..))))))...(....)..)))...........................(((((....))))).. (-28.47 = -28.80 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:33 2006