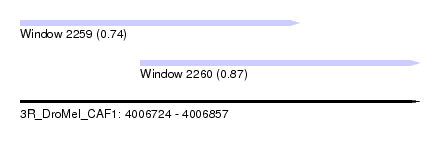
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,006,724 – 4,006,857 |
| Length | 133 |
| Max. P | 0.874155 |

| Location | 4,006,724 – 4,006,817 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 94.62 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -22.45 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
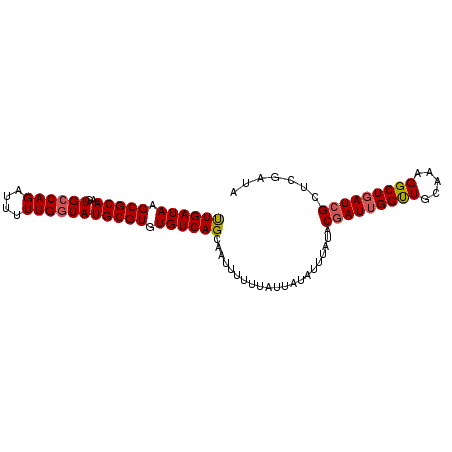
>3R_DroMel_CAF1 4006724 93 + 27905053 UUGAUAAGCGCAAGUGCCAGAUUUUUGGUAUGCGUGUGUCAGCACUUUUUUAUUAUAUUCAUACUAUUGGUUGCAAAGGCUGAUCGCUCGAUA ((((...(((((..((((((....)))))))))))((((((((.((((...((((............))))...))))))))).))))))).. ( -23.60) >DroSec_CAF1 12273 93 + 1 UUGAUAAGCGCAAGUGCCAGAUUUUUGUUAUGCGUGUGUCAGCAAUUUUUUAUUAUAUUUAUACGAUAGGUUGCAAAGGCUGAUCGCUCGAUA ......((((.....(((.(((...((.....))...))).(((((...((((..((....))..)))))))))...)))....))))..... ( -16.10) >DroSim_CAF1 12855 93 + 1 UUGAUAAGCGCAAGUGCCAGAUUUUUGGUAUGCGUGUGUCAGCAAUUUUUUAUUAUAUUUAUACGAUUGGUUGCAAAGGCUGAUCGCUCGAUA ((((((.(((((..((((((....))))))))))).)))))).....................((((..(((.....)))..))))....... ( -25.10) >DroYak_CAF1 12278 93 + 1 CUGAUAAGCGCAAGUGCCAGAUUUUUGGUAUGCGUGUGUCAGCAAAUUUGUAUUAUAUUUAUACGAUUGGCUACAAAGGCUGAUCGCUCGAUA ((((((.(((((..((((((....))))))))))).)))))).....................((((..(((.....)))..))))....... ( -29.20) >consensus UUGAUAAGCGCAAGUGCCAGAUUUUUGGUAUGCGUGUGUCAGCAAUUUUUUAUUAUAUUUAUACGAUUGGUUGCAAAGGCUGAUCGCUCGAUA ((((((.(((((..((((((....))))))))))).)))))).....................(((((((((.....)))))))))....... (-22.45 = -22.82 + 0.37)

| Location | 4,006,764 – 4,006,857 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 95.16 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -21.39 |
| Energy contribution | -22.45 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4006764 93 + 27905053 AGCACUUUUUUAUUAUAUUCAUACUAUUGGUUGCAAAGGCUGAUCGCUCGAUAAGGAUAAUGAAUUUUGCGCGCGGCUAGCGUUAAAAACUUU ............................((((...((.((((..(((.((...(((((.....))))).)).)))..)))).))...)))).. ( -17.30) >DroSec_CAF1 12313 93 + 1 AGCAAUUUUUUAUUAUAUUUAUACGAUAGGUUGCAAAGGCUGAUCGCUCGAUAAGGAUAAUGAAUUUUGCGCGCGGCUAGCGUUAAAAACUUU .((((...((((((((.(((((.(((..((((((....)).))))..)))))))).))))))))..))))((((.....)))).......... ( -23.30) >DroSim_CAF1 12895 93 + 1 AGCAAUUUUUUAUUAUAUUUAUACGAUUGGUUGCAAAGGCUGAUCGCUCGAUAAGGAUAAUGAAUUUUGCGCGCGGCUAGCGUUAAAAACUUU .((((...((((((((.(((((.((((..(((.....)))..))))....))))).))))))))..))))((((.....)))).......... ( -25.30) >DroYak_CAF1 12318 93 + 1 AGCAAAUUUGUAUUAUAUUUAUACGAUUGGCUACAAAGGCUGAUCGCUCGAUAAGGAUAAUGAAUUUUGCGCGCGGCUAGCGUUAAAAUCUUU .(((((....((((((.(((((.((((..(((.....)))..))))....))))).))))))...)))))((((.....)))).......... ( -26.20) >consensus AGCAAUUUUUUAUUAUAUUUAUACGAUUGGUUGCAAAGGCUGAUCGCUCGAUAAGGAUAAUGAAUUUUGCGCGCGGCUAGCGUUAAAAACUUU .((((...((((((((.(((((.(((((((((.....)))))))))....))))).))))))))..))))((((.....)))).......... (-21.39 = -22.45 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:23 2006