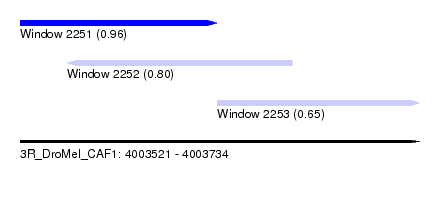
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,003,521 – 4,003,734 |
| Length | 213 |
| Max. P | 0.959958 |

| Location | 4,003,521 – 4,003,626 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4003521 105 + 27905053 AAAC---------------CCAAAAUAUAUAUAAAACCAAAGCCCAAAAGCUGCAACAACAAUGGGCUUGGGCCAGGCUAAGUGCCAAGUGAAAAUGGUUUGGGGCAUCAACUGUCAAAA ...(---------------((((.............((((.(((((................)))))))))(((.(((.....)))..((....))))))))))................ ( -26.19) >DroSec_CAF1 9122 119 + 1 AAACC-ACAAAAAAAAAAUCCAAAAUAUAUAUAAAACCAAAGCCCGAAAGCUGCAACAACAAUGGGCUUGGGCCAGGCUAAGUGCCAAGUGAAAAUGGUUUGGGGCAUCAACUGUCAAAA .....-...........................((((((..(((((..((((.((.......)))))))))))..(((.....))).........))))))..((((.....)))).... ( -23.80) >DroSim_CAF1 9700 116 + 1 AAACC-ACAAA---AAAAUCCAAAAUAUAUAUAAAACCAAAGCCCGAAAGCUGCAACAACAAUGGGCUUGGGCCAGGCUAAGUGCCAAGUGAAAAUGGUUUGGGGCAUCAACUGUCAAAA .....-.....---...................((((((..(((((..((((.((.......)))))))))))..(((.....))).........))))))..((((.....)))).... ( -23.80) >DroYak_CAF1 9096 119 + 1 AAACCAACAAA-AAAAUAAACAAAAUAUAUAUAAAACCAAAGACCAAAAGCUGCAACAACAAUGGGCUUGGGCCAGGCUAAGUGGCAAGUGAAAAUGGUUUGGGGCAUCAACUGUCAAAA ...........-........................(((..(((((...(((((....((....((((((...))))))..)).)).))).....))))))))((((.....)))).... ( -19.90) >consensus AAACC_ACAAA___AAAAUCCAAAAUAUAUAUAAAACCAAAGCCCAAAAGCUGCAACAACAAUGGGCUUGGGCCAGGCUAAGUGCCAAGUGAAAAUGGUUUGGGGCAUCAACUGUCAAAA .................................((((((..(((((..((((.((.......)))))))))))..(((.....))).........))))))..((((.....)))).... (-20.55 = -20.80 + 0.25)

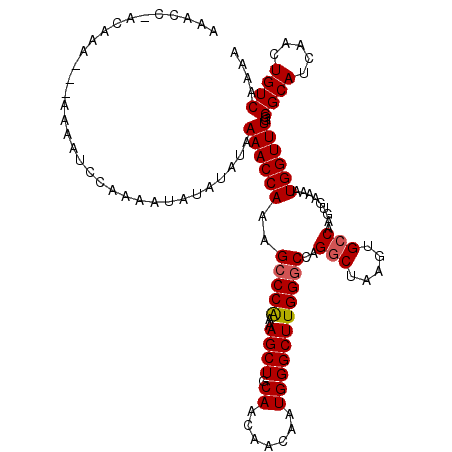
| Location | 4,003,546 – 4,003,666 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -39.03 |
| Energy contribution | -39.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4003546 120 - 27905053 GUUUUCUGGGCUGCGACGACGACUGUGACUGUGCUGUCACUUUUGACAGUUGAUGCCCCAAACCAUUUUCACUUGGCACUUAGCCUGGCCCAAGCCCAUUGUUGUUGCAGCUUUUGGGCU ((((...(((((((((((((((..((((..(((((((((....))))))(((......)))..)))..))))..(((.....))).(((....)))..)))))))))))))))..)))). ( -42.30) >DroSec_CAF1 9161 120 - 1 GUUUUCUGGGCUGCGGCGACGACUGUGACUGUGCUGUCACUUUUGACAGUUGAUGCCCCAAACCAUUUUCACUUGGCACUUAGCCUGGCCCAAGCCCAUUGUUGUUGCAGCUUUCGGGCU ((((...(((((((((((((((..((((..(((((((((....))))))(((......)))..)))..))))..(((.....))).(((....)))..)))))))))))))))..)))). ( -41.10) >DroSim_CAF1 9736 120 - 1 GUUUUCUGGGCUGCGGCGACGACUGUGACUGUGCUGUCACUUUUGACAGUUGAUGCCCCAAACCAUUUUCACUUGGCACUUAGCCUGGCCCAAGCCCAUUGUUGUUGCAGCUUUCGGGCU ((((...(((((((((((((((..((((..(((((((((....))))))(((......)))..)))..))))..(((.....))).(((....)))..)))))))))))))))..)))). ( -41.10) >DroYak_CAF1 9135 120 - 1 GUUUUCUGGGCUGCGACGACGACUGUGACUGUGCUGUCACUUUUGACAGUUGAUGCCCCAAACCAUUUUCACUUGCCACUUAGCCUGGCCCAAGCCCAUUGUUGUUGCAGCUUUUGGUCU .......(((((((((((((((..((((..(((((((((....))))))(((......)))..)))..))))..((((.......)))).........)))))))))))))))....... ( -37.30) >consensus GUUUUCUGGGCUGCGACGACGACUGUGACUGUGCUGUCACUUUUGACAGUUGAUGCCCCAAACCAUUUUCACUUGGCACUUAGCCUGGCCCAAGCCCAUUGUUGUUGCAGCUUUCGGGCU ((((...(((((((((((((((..((((..(((((((((....))))))(((......)))..)))..))))..(((.....))).(((....)))..)))))))))))))))..)))). (-39.03 = -39.27 + 0.25)

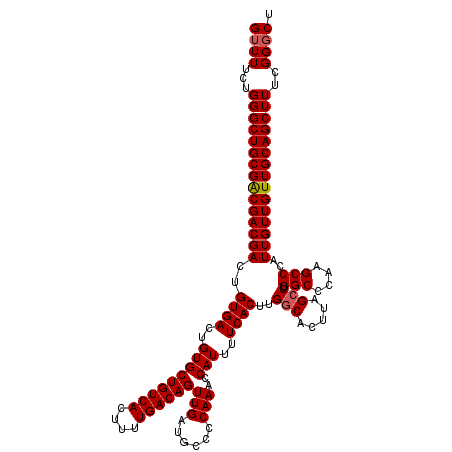
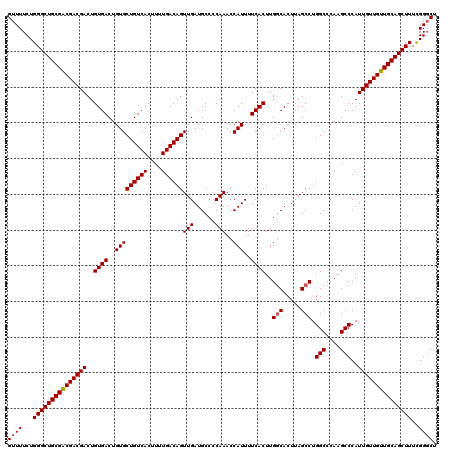
| Location | 4,003,626 – 4,003,734 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.96 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4003626 108 + 27905053 GUGACAGCACAGUCACAGUCGUCGUCGCAGCCCAGAAAACUGAAAACUGAGC------------GCCGAACACUGAAAACAGAAAGCAGAGCCAACGGUGGCUCAAAUUGAAAAUUAACU (((((......)))))..(((.(((..(((..(((....)))....))).))------------).)))...(((....)))......((((((....))))))................ ( -26.20) >DroSec_CAF1 9241 108 + 1 GUGACAGCACAGUCACAGUCGUCGCCGCAGCCCAGAAAACUGAAAACUGAGC------------GCCGAACACUGAAAACAGAAAGCAGAGCCAACGGUGGCUCAAAUUGAAAAUUAACU (((((......)))))..(((.(((..(((..(((....)))....))).))------------).)))...(((....)))......((((((....))))))................ ( -27.90) >DroSim_CAF1 9816 108 + 1 GUGACAGCACAGUCACAGUCGUCGCCGCAGCCCAGAAAACUGAAAACUGAGC------------GCCGAACACUGAAAACAGAAAGCAGAGCCAACGGUGGCUCAAAUUGAAAAUUAACU (((((......)))))..(((.(((..(((..(((....)))....))).))------------).)))...(((....)))......((((((....))))))................ ( -27.90) >DroYak_CAF1 9215 120 + 1 GUGACAGCACAGUCACAGUCGUCGUCGCAGCCCAGAAAACUGAAAACUGAACUGAACACUGAAAACAGAAAACUGAAAACAGAAAGCAGCGCCAACGGUGGCUCAAAUUGAAAAUUAACU (((((......)))))((((..((((((.((.(((....)))................(((....(((....)))....)))...)).)))...)))..))))................. ( -24.80) >consensus GUGACAGCACAGUCACAGUCGUCGCCGCAGCCCAGAAAACUGAAAACUGAGC____________GCCGAACACUGAAAACAGAAAGCAGAGCCAACGGUGGCUCAAAUUGAAAAUUAACU (((((......)))))(((..((...((....(((....)))..............................(((....)))...)).((((((....)))))).....))..))).... (-21.23 = -21.47 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:16 2006