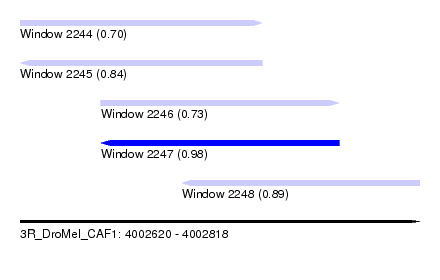
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,002,620 – 4,002,818 |
| Length | 198 |
| Max. P | 0.977841 |

| Location | 4,002,620 – 4,002,740 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.03 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4002620 120 + 27905053 AGUCACAAGACAAAAGUUAUUGCCAAAAUUUUCGGCAUGUUUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCUUGGGCUAUCAUAAAAAUUUCCGACUCACGA .(((....)))((((((...((((.........)))).....))))))(((((....)))))....((((.((((..((((((((((((....))...))))))))))..))))..)))) ( -28.00) >DroSec_CAF1 8212 120 + 1 AGUCACAAGACAAAAGUUAUUGCCAAAAUUUUCGGCAUGUUUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCCUGGGCUAUCAUAAAAAUUUCCAACUCACGA ........(((((((((...((((.........)))).....)))).((((((....))))))......)))))(((((((((((((((....))...))))))))...)))))...... ( -25.60) >DroSim_CAF1 8783 120 + 1 AGUCACAAGACAAAAGUUAUUGCCAAAAUUUUCGGCAUGUUUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCCUGGGCUAUCAUAAAAAUUUCCGACUCACGA .(((....)))((((((...((((.........)))).....))))))(((((....)))))....((((.((((..((((((((((((....))...))))))))))..))))..)))) ( -29.10) >DroYak_CAF1 8142 120 + 1 AGUCACAAGACAAAAGUUAUUGCCAAAAUUUUCGGCAUGUUUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCUUGGGCUAUCAUAAAAAUUUCCGACUCACGA .(((....)))((((((...((((.........)))).....))))))(((((....)))))....((((.((((..((((((((((((....))...))))))))))..))))..)))) ( -28.00) >consensus AGUCACAAGACAAAAGUUAUUGCCAAAAUUUUCGGCAUGUUUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCCUGGGCUAUCAUAAAAAUUUCCGACUCACGA .(((....)))((((((...((((.........)))).....))))))(((((....)))))....((((.((((..((((((((((((....))...))))))))))..))))..)))) (-27.10 = -27.35 + 0.25)

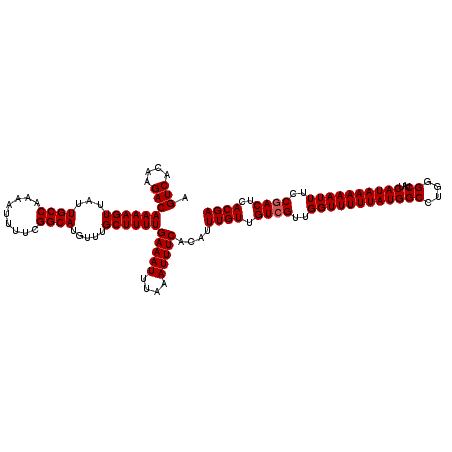
| Location | 4,002,620 – 4,002,740 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.03 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -24.91 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4002620 120 - 27905053 UCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAAACAUGCCGAAAAUUUUGGCAAUAACUUUUGUCUUGUGACU (((..(((((....((((((((...((....))))))))))....)))).(((((.(((((((....)))))...........((((((.....))))))...)).))))).)..))).. ( -24.30) >DroSec_CAF1 8212 120 - 1 UCGUGAGUUGGAAAUUUUUAUGAUAGCCCAGGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAAACAUGCCGAAAAUUUUGGCAAUAACUUUUGUCUUGUGACU (((..((((((...((((((((...((....)))))))))))))))(((((......((((((....))))))..........((((((.....)))))).......))))))..))).. ( -28.50) >DroSim_CAF1 8783 120 - 1 UCGUGAGUCGGAAAUUUUUAUGAUAGCCCAGGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAAACAUGCCGAAAAUUUUGGCAAUAACUUUUGUCUUGUGACU (((..(((((....((((((((...((....))))))))))....)))).(((((.(((((((....)))))...........((((((.....))))))...)).))))).)..))).. ( -26.20) >DroYak_CAF1 8142 120 - 1 UCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAAACAUGCCGAAAAUUUUGGCAAUAACUUUUGUCUUGUGACU (((..(((((....((((((((...((....))))))))))....)))).(((((.(((((((....)))))...........((((((.....))))))...)).))))).)..))).. ( -24.30) >consensus UCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAAACAUGCCGAAAAUUUUGGCAAUAACUUUUGUCUUGUGACU (((..(((((....((((((((...((....))))))))))....)))).(((((.(((((((....)))))...........((((((.....))))))...)).))))).)..))).. (-24.91 = -24.72 + -0.19)

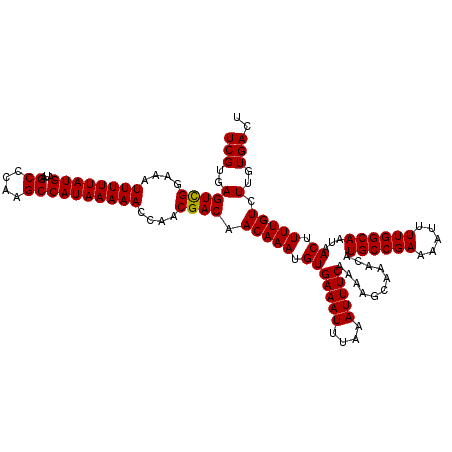
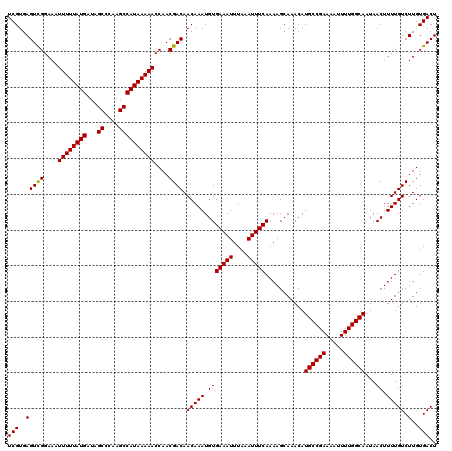
| Location | 4,002,660 – 4,002,778 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4002660 118 + 27905053 UUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCUUGGGCUAUCAUAAAAAUUUCCGACUCACGAGCAUGUGAGCUCUUAAGUGUG--UGUGCUGUUUUGGGCCA ..((((..(((((....((..((((((((..((((..((((((((((((....))...))))))))))..))))....((((......)))).))))))))--.))...))))))))).. ( -30.70) >DroSec_CAF1 8252 118 + 1 UUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCCUGGGCUAUCAUAAAAAUUUCCAACUCACGAGCAUGUGAGCUCCUAAGUAUG--UGUUUUGUUUUGGGCCA ..(((..((((((....))))))(((.((((((.(((((((((((((((....))...))))))))...)))))..)).)))).)))))).((((((....--........))))))... ( -24.00) >DroSim_CAF1 8823 118 + 1 UUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCCUGGGCUAUCAUAAAAAUUUCCGACUCACGAGCAUGUGAGCUCUUAAGUGUG--UGUGUUGUUUUGGGCCA ..((((..(((((....((..((((((((..((((..((((((((((((....))...))))))))))..))))....((((......)))).))))))))--.))...))))))))).. ( -31.80) >DroYak_CAF1 8182 120 + 1 UUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCUUGGGCUAUCAUAAAAAUUUCCGACUCACGAGCAUGUGAGCUCUUAAGUGUGUGUGUGUUGUUUUGGGCCA ..((((..(((((........((((((((..((((..((((((((((((....))...))))))))))..))))....((((......)))).))))))))........))))))))).. ( -31.29) >consensus UUGCUUUUGAAAUUUAAAUUUCACAUUUGUUGUCGUUGGUUUUUAUGGCCUGGGCUAUCAUAAAAAUUUCCGACUCACGAGCAUGUGAGCUCUUAAGUGUG__UGUGUUGUUUUGGGCCA ..((((..(((((........((((((((..((((..((((((((((((....))...))))))))))..))))....((((......)))).))))))))........))))))))).. (-28.42 = -28.92 + 0.50)

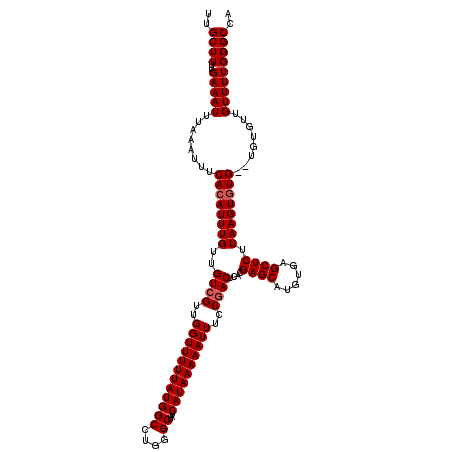
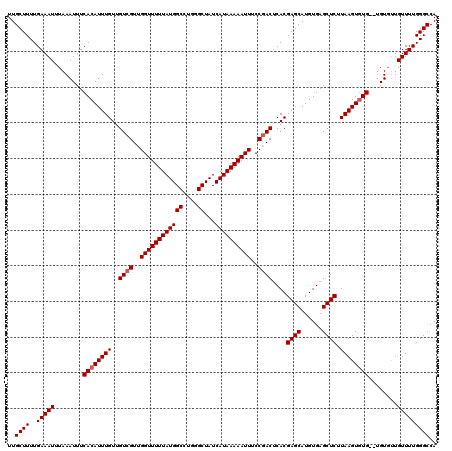
| Location | 4,002,660 – 4,002,778 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -22.16 |
| Energy contribution | -21.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4002660 118 - 27905053 UGGCCCAAAACAGCACA--CACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAA ............((...--.........((((......))))((..((((....((((((((...((....))))))))))....)))).)).....((((((....))))))...)).. ( -22.40) >DroSec_CAF1 8252 118 - 1 UGGCCCAAAACAAAACA--CAUACUUAGGAGCUCACAUGCUCGUGAGUUGGAAAUUUUUAUGAUAGCCCAGGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAA ...............((--(((..(((.((((......)))).)))(((((...((((((((...((....)))))))))))))))........)))))..................... ( -25.50) >DroSim_CAF1 8823 118 - 1 UGGCCCAAAACAACACA--CACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAGGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAA ..((.....(((.....--.........((((......))))((..((((....((((((((...((....))))))))))....)))).))...)))(((((....)))))....)).. ( -23.50) >DroYak_CAF1 8182 120 - 1 UGGCCCAAAACAACACACACACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAA ..((.....(((................((((......))))((..((((....((((((((...((....))))))))))....)))).))...)))(((((....)))))....)).. ( -21.60) >consensus UGGCCCAAAACAACACA__CACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAAACCAACGACAACAAAUGUGAAAUUUAAAUUUCAAAAGCAA ..((.....(((................((((......))))((..((((....((((((((...((....))))))))))....)))).))...)))(((((....)))))....)).. (-22.16 = -21.98 + -0.19)

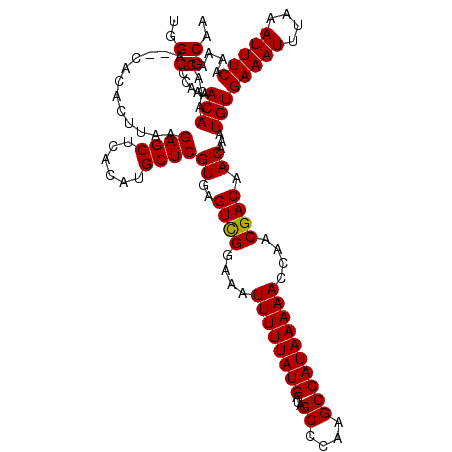

| Location | 4,002,700 – 4,002,818 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.68 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -26.56 |
| Energy contribution | -27.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4002700 118 - 27905053 UAGUUUUUUGGCUUAAACGCAGAGGCCAAUGCAGUUUUAAUGGCCCAAAACAGCACA--CACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAA ...((((.((((((....((.(.(((((.((......)).))))))...........--..((((((.((((......)))).))))).)...............))..)))))).)))) ( -27.40) >DroSec_CAF1 8292 118 - 1 UAGUUUUUUGGCUUAAACGAUGAGGCCAAUGCAGUUUUAAUGGCCCAAAACAAAACA--CAUACUUAGGAGCUCACAUGCUCGUGAGUUGGAAAUUUUUAUGAUAGCCCAGGCCAUAAAA ..((...(((((((........))))))).)).......((((((............--...(((((.((((......)))).))))).((..(((.....)))..))..)))))).... ( -32.10) >DroSim_CAF1 8863 118 - 1 UAGUUUUUUGGCUUAAACGCUGAGGCCAAUGCAGUUUUAAUGGCCCAAAACAACACA--CACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAGGCCAUAAAA ..((...(((((((........))))))).)).......((((((............--...(((((.((((......)))).))))).((..(((.....)))..))..)))))).... ( -30.40) >DroYak_CAF1 8222 120 - 1 UAGUUUUUUGGCUUAAACGCAGAACCCAAUGCAGUUUUAAUGGCCCAAAACAACACACACACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAA ..(((((..((((.(((((((........))).))))....)))).)))))..........((((((.((((......)))).))))).).....(((((((...((....))))))))) ( -25.50) >consensus UAGUUUUUUGGCUUAAACGCAGAGGCCAAUGCAGUUUUAAUGGCCCAAAACAACACA__CACACUUAAGAGCUCACAUGCUCGUGAGUCGGAAAUUUUUAUGAUAGCCCAAGCCAUAAAA ..((...(((((((........))))))).)).......((((((.................(((((.((((......)))).))))).((..(((.....)))..))..)))))).... (-26.56 = -27.13 + 0.56)

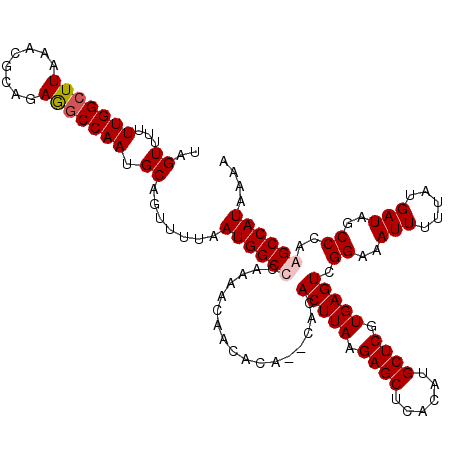
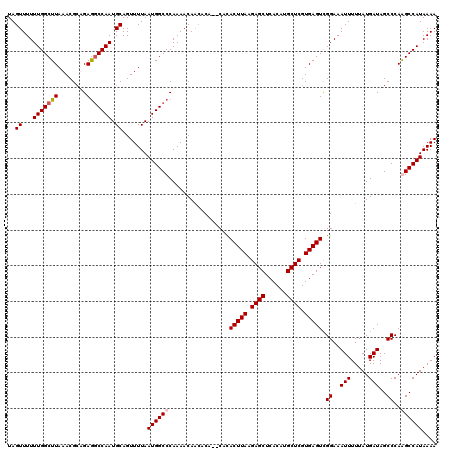
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:11 2006