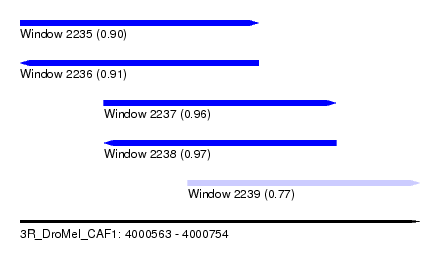
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,000,563 – 4,000,754 |
| Length | 191 |
| Max. P | 0.966437 |

| Location | 4,000,563 – 4,000,677 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -32.65 |
| Energy contribution | -33.43 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4000563 114 + 27905053 GGAAUGCUGCUGAUGCAGUUGCUGCCGUAGCCGCUGCAGCGGACAAUGCACAUUGUGGCGGCACCUUUGGCACUGCGGCAUGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA (((..((((((..((((((((((((((..(((((..(((((.....)))....))..))))).....)))))..))))).)))).))))))...(.....).)))......... ( -44.10) >DroSec_CAF1 6209 94 + 1 GGAAUGCUGUUG---------CU------GCUGCUGCAGCGGACAAUGCACAUUGUGGCGGCACCUUUGGCUCUGCGGCAUGC-----AGCCAAGAGCAACAUCCACAGCAACA ....((((((((---------((------((..(((((........))))....)..))))))......(((((..(((....-----.))).))))).......))))))... ( -36.70) >DroSim_CAF1 6268 96 + 1 GGAAUGCUG------------CU------GCUGCUGCAGCGGACAAUGCACAUUGUGGCGGCACCUUUGGCUCUGCGGCAUGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA ....(((((------------.(------(((((..(((((.....)))....))..))))))......(((((..(((.(((...)))))).)))))........)))))... ( -36.70) >consensus GGAAUGCUG_UG_________CU______GCUGCUGCAGCGGACAAUGCACAUUGUGGCGGCACCUUUGGCUCUGCGGCAUGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA ....(((((....................(((((..(((((.....)))....))..))))).......(((((..(((.(((...)))))).)))))........)))))... (-32.65 = -33.43 + 0.78)

| Location | 4,000,563 – 4,000,677 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -31.29 |
| Energy contribution | -32.07 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4000563 114 - 27905053 UGUUGCUGUGGAUGUUGCUCUUGGCUGCUUUGCAUGCCGCAGUGCCAAAGGUGCCGCCACAAUGUGCAUUGUCCGCUGCAGCGGCUACGGCAGCAACUGCAUCAGCAGCAUUCC ((((((((((.(.((((((((((((((((..(((.((.(((((((((...(((....)))..)).)))))).).))))))))))))).)).))))))).)).)))))))).... ( -50.30) >DroSec_CAF1 6209 94 - 1 UGUUGCUGUGGAUGUUGCUCUUGGCU-----GCAUGCCGCAGAGCCAAAGGUGCCGCCACAAUGUGCAUUGUCCGCUGCAGCAGC------AG---------CAACAGCAUUCC ...((((((((((...(((((((((.-----....)))).)))))....(((((((......)).)))))))))(((((....))------))---------).)))))).... ( -37.90) >DroSim_CAF1 6268 96 - 1 UGUUGCUGUGGAUGUUGCUCUUGGCUGCUUUGCAUGCCGCAGAGCCAAAGGUGCCGCCACAAUGUGCAUUGUCCGCUGCAGCAGC------AG------------CAGCAUUCC (((((((((((((..(((.(.((((.(((((((.....)))))))....(....)))))....).)))..))))((....)).))------))------------))))).... ( -38.80) >consensus UGUUGCUGUGGAUGUUGCUCUUGGCUGCUUUGCAUGCCGCAGAGCCAAAGGUGCCGCCACAAUGUGCAUUGUCCGCUGCAGCAGC______AG_________CA_CAGCAUUCC ((((((.((((((..(((.(((((((...((((.....)))))))))).((.....)).....).)))..)))))).))))))............................... (-31.29 = -32.07 + 0.78)

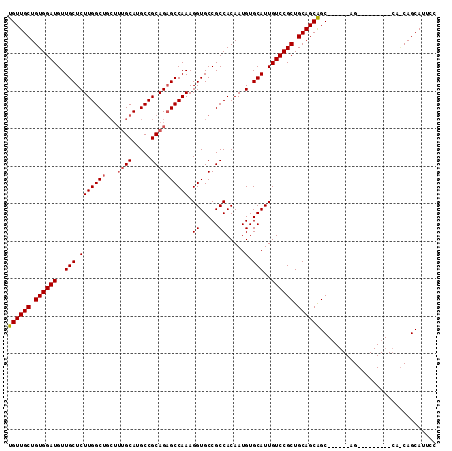
| Location | 4,000,603 – 4,000,714 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -36.95 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4000603 111 + 27905053 GGACAAUGCACAUUGUGGCGGCACCUUUGGCACUGCGGCAUGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA---------ACUGCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAA ...((((...((..(((....)))...))((((((((((.(((...))))))...(((((((......(((...---------..)))......))))))))))))))......)))).. ( -34.60) >DroSec_CAF1 6234 103 + 1 GGACAAUGCACAUUGUGGCGGCACCUUUGGCUCUGCGGCAUGC-----AGCCAAGAGCAACAUCCACAGCAACA------------GCAAGAACUGUUGUUGCAGUGCAAAUUAAUUGAA ......(((((..(((((.(....)....(((((..(((....-----.))).))))).....)))))((((((------------(((.....))))))))).)))))........... ( -38.20) >DroSim_CAF1 6290 108 + 1 GGACAAUGCACAUUGUGGCGGCACCUUUGGCUCUGCGGCAUGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA------------GCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAA ......(((((...((((.(....)....(((((..(((.(((...)))))).))))).....))))(((((((------------(......))))))))...)))))........... ( -37.90) >DroYak_CAF1 6073 120 + 1 GGACAAUGCACAUUGUGGCGGCACCUUUGGCUCUGCGGCAUGCAAAGCAGCCAAGAGCAACAUCCACAGCAACAUCCAGAGGAACGGCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAA ......(((((..((..((((((..(((.(((((..(((.(((...)))))).)))))..........((....(((...)))...)).)))..))))))..)))))))........... ( -37.10) >consensus GGACAAUGCACAUUGUGGCGGCACCUUUGGCUCUGCGGCAUGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA____________GCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAA ......(((((..((..((((((..(((.(((((..(((.(((...)))))).)))))..........((................)).)))..))))))..)))))))........... (-30.10 = -30.67 + 0.56)

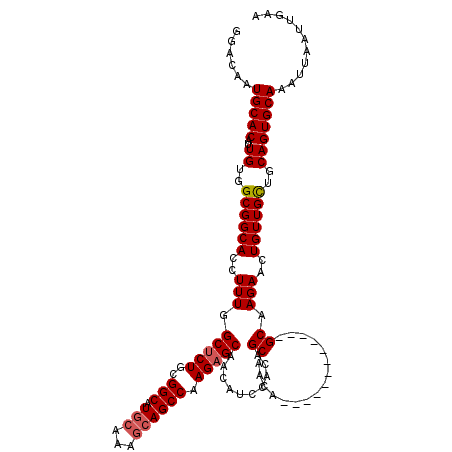
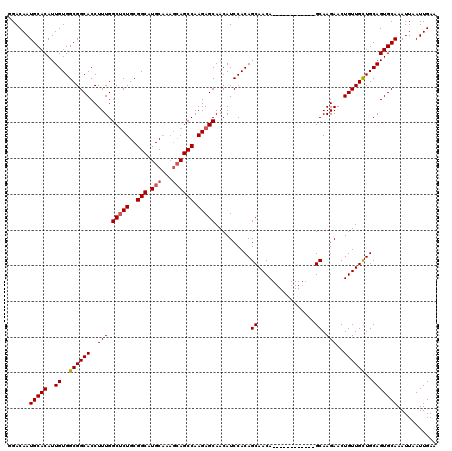
| Location | 4,000,603 – 4,000,714 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -39.09 |
| Consensus MFE | -32.21 |
| Energy contribution | -32.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4000603 111 - 27905053 UUCAAUUAAUUUGCACUGCAGCAACAGUUCUUGCAGU---------UGUUGCUGUGGAUGUUGCUCUUGGCUGCUUUGCAUGCCGCAGUGCCAAAGGUGCCGCCACAAUGUGCAUUGUCC ...........(((((.((((((((.((....)).))---------))))))(((((.((..((..(((((....((((.....)))).)))))..))..)))))))..)))))...... ( -40.50) >DroSec_CAF1 6234 103 - 1 UUCAAUUAAUUUGCACUGCAACAACAGUUCUUGC------------UGUUGCUGUGGAUGUUGCUCUUGGCU-----GCAUGCCGCAGAGCCAAAGGUGCCGCCACAAUGUGCAUUGUCC ...........(((((.....(((((((....))------------))))).(((((.((..((..((((((-----((.....))..))))))..))..)))))))..)))))...... ( -35.60) >DroSim_CAF1 6290 108 - 1 UUCAAUUAAUUUGCACUGCAGCAACAGUUCUUGC------------UGUUGCUGUGGAUGUUGCUCUUGGCUGCUUUGCAUGCCGCAGAGCCAAAGGUGCCGCCACAAUGUGCAUUGUCC ...........(((((.(((((((((((....))------------)))))))))((.((..((..((((((...((((.....))))))))))..))..)))).....)))))...... ( -41.90) >DroYak_CAF1 6073 120 - 1 UUCAAUUAAUUUGCACUGCAGCAACAGUUCUUGCCGUUCCUCUGGAUGUUGCUGUGGAUGUUGCUCUUGGCUGCUUUGCAUGCCGCAGAGCCAAAGGUGCCGCCACAAUGUGCAUUGUCC ...........(((((.((((((.(((..............)))..))))))(((((.((..((..((((((...((((.....))))))))))..))..)))))))..)))))...... ( -38.34) >consensus UUCAAUUAAUUUGCACUGCAGCAACAGUUCUUGC____________UGUUGCUGUGGAUGUUGCUCUUGGCUGCUUUGCAUGCCGCAGAGCCAAAGGUGCCGCCACAAUGUGCAUUGUCC ...........(((((.((((((.......................))))))(((((.((..((..((((((...((((.....))))))))))..))..)))))))..)))))...... (-32.21 = -32.77 + 0.56)

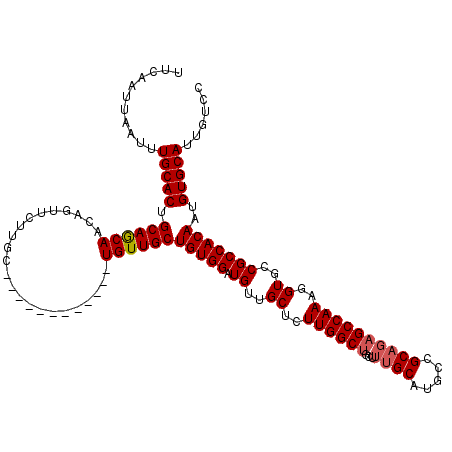
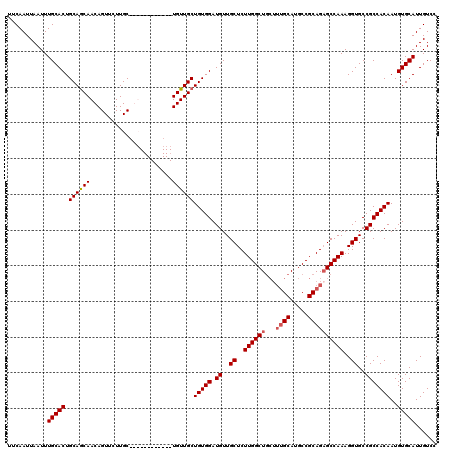
| Location | 4,000,643 – 4,000,754 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.59 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -23.10 |
| Energy contribution | -22.91 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4000643 111 + 27905053 UGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA---------ACUGCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAAGCAUCGCAAGGCAAUGUCAACGACAGCAACAUCACGUGUU (((......(((...(((((((......(((...---------..)))......)))))))((.((((............)))).))..)))..((((...)))))))............ ( -26.70) >DroSec_CAF1 6274 103 + 1 UGC-----AGCCAAGAGCAACAUCCACAGCAACA------------GCAAGAACUGUUGUUGCAGUGCAAAUUAAUUGAAGCAUCGCAAGGCAAUGUCAACGACAGCAACAUCACGUGUU (((-----.(((..((((......(((.((((((------------(((.....))))))))).)))(((.....)))..)).))....)))..((((...)))))))............ ( -26.50) >DroSim_CAF1 6330 108 + 1 UGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA------------GCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAAGCAUCGCAAGGCAAUGUCAACGACAGCAACAUCACGUGUU (((......(((..((((........((((((((------------(......)))))))))((((........))))..)).))....)))..((((...)))))))............ ( -27.60) >DroYak_CAF1 6113 120 + 1 UGCAAAGCAGCCAAGAGCAACAUCCACAGCAACAUCCAGAGGAACGGCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAAGCAUCGCAAGGCAAUGUCAACGACAGCAACAUCACGUGUU (((......(((...(((((((......((....(((...)))...))......)))))))((.((((............)))).))..)))..((((...)))))))............ ( -27.40) >consensus UGCAAAGCAGCCAAGAGCAACAUCCACAGCAACA____________GCAAGAACUGUUGCUGCAGUGCAAAUUAAUUGAAGCAUCGCAAGGCAAUGUCAACGACAGCAACAUCACGUGUU (((......(((...(((((((......((................))......)))))))((.((((............)))).))..)))..((((...)))))))............ (-23.10 = -22.91 + -0.19)

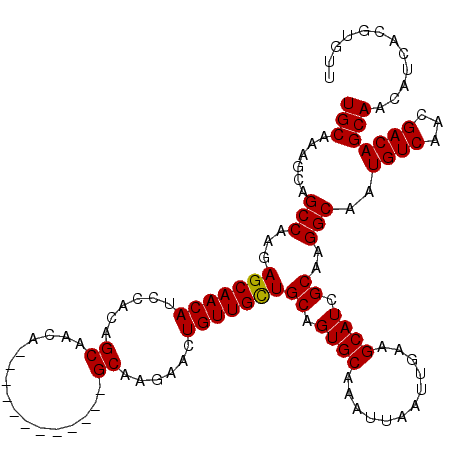

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:03 2006