
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,996,103 – 3,996,223 |
| Length | 120 |
| Max. P | 0.930393 |

| Location | 3,996,103 – 3,996,223 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -25.92 |
| Energy contribution | -25.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3996103 120 + 27905053 CUCUCUUUUGGCCCGUUCUCCUAUCUCUCUCUCUCUCUCUUGGACAUUUCUAUGGCGUUUUAAAAGCGUUUGACAGCGCUCAACGCUCUCAGCGCCUGUCCAUCUAAGCCAGAUCCAAAA ......((((((..(....)....................((((((.......((((((.....((((((.((......))))))))...)))))))))))).....))))))....... ( -25.71) >DroSec_CAF1 1858 120 + 1 CUCUCUUUUGGCCCGUUCUCCUCUCUCACUCCCUCUCUCUUGGACAUUUCUAUGGCGUUUUAAAAGCGUUUGACAGCGCUCAACGCUCUCAGCGCCUGUCCAUCUAAGCCAGAUCCGAAA .((...((((((..((...........))...........((((((.......((((((.....((((((.((......))))))))...)))))))))))).....))))))...)).. ( -26.41) >DroSim_CAF1 1866 120 + 1 CUCUCUUUUGGCCCGUUCUCCUCUCGCACUCCCUCUCUCUUGGACAUUUCUAUGGCGUUUUAAAAGCGUUUGACAGCGCUCAACGCUCUCAGCGCCUGUCCAUCUAAGCCAGAUCCGAAA .((...((((((.((.........))..............((((((.......((((((.....((((((.((......))))))))...)))))))))))).....))))))...)).. ( -27.41) >DroYak_CAF1 1332 108 + 1 CUCUCUUUUGGCCCGCUCGCC------------UCUCUCUUGGACAUUUCUGUGGCGUUUUAAAAGCGUUUGACAGCGCUCAACGCUCUCAGCGCCUGUCCAUCUAAGCCAGGUUCGAAA ......((((..((....((.------------.......((((((...(((.((((((.....(((((......))))).))))))..)))....)))))).....))..))..)))). ( -28.12) >consensus CUCUCUUUUGGCCCGUUCUCCUCUCUCACUCCCUCUCUCUUGGACAUUUCUAUGGCGUUUUAAAAGCGUUUGACAGCGCUCAACGCUCUCAGCGCCUGUCCAUCUAAGCCAGAUCCGAAA ......((((((..(....)....................((((((.......((((((.....((((((.((......))))))))...)))))))))))).....))))))....... (-25.92 = -25.73 + -0.19)

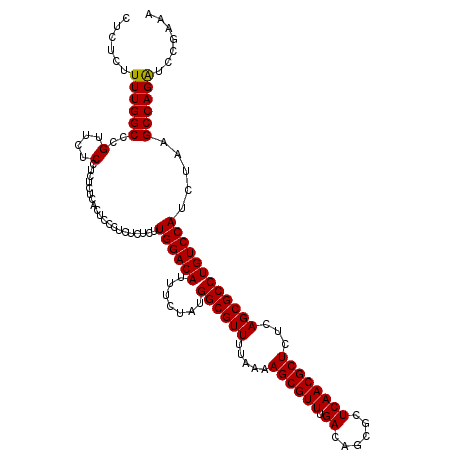

| Location | 3,996,103 – 3,996,223 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -31.15 |
| Energy contribution | -30.96 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3996103 120 - 27905053 UUUUGGAUCUGGCUUAGAUGGACAGGCGCUGAGAGCGUUGAGCGCUGUCAAACGCUUUUAAAACGCCAUAGAAAUGUCCAAGAGAGAGAGAGAGAGAUAGGAGAACGGGCCAAAAGAGAG ((((((.(((..(((...((((((((((.((((((((((((......)).))))))))))...)))).......))))))...)))..)))........(.....)...))))))..... ( -32.21) >DroSec_CAF1 1858 120 - 1 UUUCGGAUCUGGCUUAGAUGGACAGGCGCUGAGAGCGUUGAGCGCUGUCAAACGCUUUUAAAACGCCAUAGAAAUGUCCAAGAGAGAGGGAGUGAGAGAGGAGAACGGGCCAAAAGAGAG (((((..(((..(((...((((((((((.((((((((((((......)).))))))))))...)))).......))))))...)))..))).)))))..((........))......... ( -32.41) >DroSim_CAF1 1866 120 - 1 UUUCGGAUCUGGCUUAGAUGGACAGGCGCUGAGAGCGUUGAGCGCUGUCAAACGCUUUUAAAACGCCAUAGAAAUGUCCAAGAGAGAGGGAGUGCGAGAGGAGAACGGGCCAAAAGAGAG (((((.((((..(((...((((((((((.((((((((((((......)).))))))))))...)))).......))))))...)))..))).).)))))((........))......... ( -34.01) >DroYak_CAF1 1332 108 - 1 UUUCGAACCUGGCUUAGAUGGACAGGCGCUGAGAGCGUUGAGCGCUGUCAAACGCUUUUAAAACGCCACAGAAAUGUCCAAGAGAGA------------GGCGAGCGGGCCAAAAGAGAG ((((.....((((((.....(((((.((((.((....)).)))))))))...(((((((........(((....))).......)))------------))))...))))))...)))). ( -32.76) >consensus UUUCGGAUCUGGCUUAGAUGGACAGGCGCUGAGAGCGUUGAGCGCUGUCAAACGCUUUUAAAACGCCAUAGAAAUGUCCAAGAGAGAGGGAGUGAGAGAGGAGAACGGGCCAAAAGAGAG ((((.....((((((...((((((((((.((((((((((((......)).))))))))))...)))).......))))))...................(.....)))))))...)))). (-31.15 = -30.96 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:43 2006