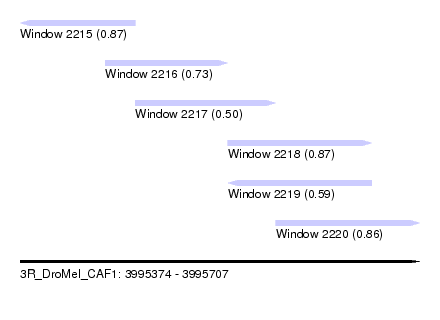
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,995,374 – 3,995,707 |
| Length | 333 |
| Max. P | 0.873813 |

| Location | 3,995,374 – 3,995,470 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 99.31 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -31.49 |
| Energy contribution | -31.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995374 96 - 27905053 AGCGAUUCGAACCGAAUCGAUCCGAUGACGACUUCAUAUAUGGAGGAUCUUUGUGUCGUUGGUGCAAAAGUGUGAACUGGGCACAGCAGAUCGGGA ..(((((((...))))))).(((........(((((....)))))(((((((((((((((.(..(....)..).)))..))))))).)))))))). ( -31.80) >DroSec_CAF1 1169 96 - 1 AGCGAUUCGAACCGAAUCGAUCCGAUGACGACUUCAUAUAUGGGGGAUCUUUGUGUCGUUGGUGCAAAAGUGUGAACUGGGCACAGCAGAUCGGGA ..(((((((...))))))).(((........(((((....)))))(((((((((((((((.(..(....)..).)))..))))))).)))))))). ( -31.00) >DroSim_CAF1 1168 96 - 1 AGCGAUUCGAACCGAAUCGAUCCGAUGACGACUUCAUAUAUGGGGGAUCUUUGUGUCGUUGGUGCAAAAGUGUGAACUGGGCACAGCAGAUCGGGA ..(((((((...))))))).(((........(((((....)))))(((((((((((((((.(..(....)..).)))..))))))).)))))))). ( -31.00) >consensus AGCGAUUCGAACCGAAUCGAUCCGAUGACGACUUCAUAUAUGGGGGAUCUUUGUGUCGUUGGUGCAAAAGUGUGAACUGGGCACAGCAGAUCGGGA ..(((((((...))))))).(((........(((((....)))))(((((((((((((((.(..(....)..).)))..))))))).)))))))). (-31.49 = -31.27 + -0.22)


| Location | 3,995,445 – 3,995,547 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 99.35 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -28.17 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995445 102 + 27905053 UCGGAUCGAUUCGGUUCGAAUCGCUUGUAUGUAAUUUGCCAAGAUCGAGUGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCGGAGGAUGGUUGCAUC ((.((((((((((...)))))))....(((((((((((.((.((((.((((.........)))))))).)).)))))))))))))).)).((((....)))) ( -26.90) >DroSec_CAF1 1240 102 + 1 UCGGAUCGAUUCGGUUCGAAUCGCUUGUAUGUAAUUUGCCAAGAUCGAGGGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCGGAGGAUGGUUGCAUC ((.((((((((((...)))))))....(((((((((((.((.((((((.((........)).)))))).)).)))))))))))))).)).((((....)))) ( -29.80) >DroSim_CAF1 1239 102 + 1 UCGGAUCGAUUCGGUUCGAAUCGCUUGUAUGUAAUUUGCCAAGAUCGAGGGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCGGAGGAUGGUUGCAUC ((.((((((((((...)))))))....(((((((((((.((.((((((.((........)).)))))).)).)))))))))))))).)).((((....)))) ( -29.80) >consensus UCGGAUCGAUUCGGUUCGAAUCGCUUGUAUGUAAUUUGCCAAGAUCGAGGGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCGGAGGAUGGUUGCAUC ((.((((((((((...)))))))....(((((((((((.((.((((((.((........)).)))))).)).)))))))))))))).)).((((....)))) (-28.17 = -28.50 + 0.33)



| Location | 3,995,470 – 3,995,587 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.62 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -27.27 |
| Energy contribution | -27.15 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995470 117 + 27905053 UGUAUGUAAUUUGCCAAGAUCGAGUGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCG---GAGGAUGGUUGCAUCUUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUA ..(((((((((((.((.((((.((((.........)))))))).)).)))))))))))...(---(((((((....)))))))).((.(((((.((......))..))))).))...... ( -30.20) >DroSec_CAF1 1265 117 + 1 UGUAUGUAAUUUGCCAAGAUCGAGGGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCG---GAGGAUGGUUGCAUCUUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUA ..(((((((((((.((.((((((.((........)).)))))).)).)))))))))))...(---(((((((....)))))))).((.(((((.((......))..))))).))...... ( -33.10) >DroSim_CAF1 1264 117 + 1 UGUAUGUAAUUUGCCAAGAUCGAGGGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCG---GAGGAUGGUUGCAUCUUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUA ..(((((((((((.((.((((((.((........)).)))))).)).)))))))))))...(---(((((((....)))))))).((.(((((.((......))..))))).))...... ( -33.10) >DroYak_CAF1 712 120 + 1 UGUAUGUAAUUUGGCAAGAUCGAGGGAAAUAAUACCAUUGGUCAUGCUAAAUUAUAUAGUCGAAGGAGGAUGGUUGCAUCUUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGUUCUUUA ..((((((((((((((.((((((.((........)).)))))).))))))))))))))((((..((((((((....))))))))....(((((......)))))..)))).......... ( -37.70) >consensus UGUAUGUAAUUUGCCAAGAUCGAGGGAAAUAAUACCAUUGGUCAUGCCAAAUUAUAUAGUCG___GAGGAUGGUUGCAUCUUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUA ..(((((((((((.((.((((((.((........)).)))))).)).))))))))))).......(((((((....)))))))..((.(((((.((......))..))))).))...... (-27.27 = -27.15 + -0.12)


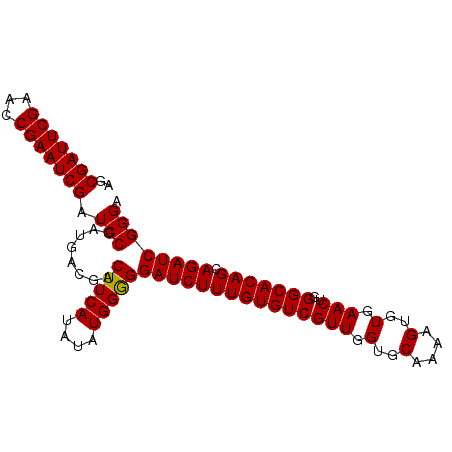
| Location | 3,995,547 – 3,995,667 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -33.01 |
| Energy contribution | -33.58 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995547 120 + 27905053 UUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUAAUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGUGGGCUGUUAAUGUUAAUCCCAAGUUAU ...(((((((((((((.(((.......)))..(((((...((((((.(((((((........)))))))))))))))))))))))))......))))))..................... ( -38.90) >DroSec_CAF1 1342 120 + 1 UUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUAAUUGGCAUGACGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGUAUGCUGUUAAUGUUAAUCCCAAGUUAU .....((.(((((.((......))..))))).))((((..((((((.((.((((........)))).))))))))((..((((((.((.(((....))).)).))))))..))))))... ( -26.30) >DroSim_CAF1 1341 120 + 1 UUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUAAUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGCGGGCUGUUAAUGUUAAUCCCAAGUUAU ...(((((((((((((.(((.......)))..(((((...((((((.(((((((........))))))))))))))))))))))(....))))))))))..................... ( -41.30) >DroYak_CAF1 792 119 + 1 UUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGUUCUUUAAUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGCGGGCUGUUAAUGUUAAUCC-AAGCUAU ...((((((((((((((((((((((.....))))).....((((((.(((((((........)))))))))))))))).)))))(....)))))))))).............-....... ( -40.20) >consensus UUUCAGCCCGUUGCCAAAUCAAUGUCCGAUGCGCACUUUAAUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGCGGGCUGUUAAUGUUAAUCCCAAGUUAU ...(((((((((((((((((.......)))...((((...((((((.(((((((........))))))))))))))))))))))))......)))))))..................... (-33.01 = -33.58 + 0.56)


| Location | 3,995,547 – 3,995,667 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.52 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995547 120 - 27905053 AUAACUUGGGAUUAACAUUAACAGCCCACUGAUAUCGCCAACACCAUCGGCUGGCAAAUUGAAAGUUUCGCCAUGCCAAUUAAAGUGCGCAUCGGACAUUGAUUUGGCAACGGGCUGAAA .....................((((((...(....)((((........(((((((((((.....)))).)))).)))((((((.((.((...)).)).))))))))))...))))))... ( -34.30) >DroSec_CAF1 1342 120 - 1 AUAACUUGGGAUUAACAUUAACAGCAUACUGAUAUCGCCAACACCAUCGGCUGGCAAAUUGAAAGUUUCGUCAUGCCAAUUAAAGUGCGCAUCGGACAUUGAUUUGGCAACGGGCUGAAA .....................((((...........((((.(......)..))))............((((..((((((((((.((.((...)).)).)))).))))))))))))))... ( -25.70) >DroSim_CAF1 1341 120 - 1 AUAACUUGGGAUUAACAUUAACAGCCCGCUGAUAUCGCCAACACCAUCGGCUGGCAAAUUGAAAGUUUCGCCAUGCCAAUUAAAGUGCGCAUCGGACAUUGAUUUGGCAACGGGCUGAAA .....................(((((((..(....)((((........(((((((((((.....)))).)))).)))((((((.((.((...)).)).))))))))))..)))))))... ( -35.60) >DroYak_CAF1 792 119 - 1 AUAGCUU-GGAUUAACAUUAACAGCCCGCUGAUAUCGCCAACACCAUCGGCUGGCAAAUUGAAAGUUUCGCCAUGCCAAUUAAAGAACGCAUCGGACAUUGAUUUGGCAACGGGCUGAAA .......-.............(((((((..(....)((((........(((((((((((.....)))).)))).)))((((((.(..((...))..).))))))))))..)))))))... ( -32.90) >consensus AUAACUUGGGAUUAACAUUAACAGCCCACUGAUAUCGCCAACACCAUCGGCUGGCAAAUUGAAAGUUUCGCCAUGCCAAUUAAAGUGCGCAUCGGACAUUGAUUUGGCAACGGGCUGAAA .....................((((((...(....)((((........(((((((((((.....)))).)))).)))((((((.((.((...)).)).))))))))))...))))))... (-30.40 = -30.52 + 0.13)



| Location | 3,995,587 – 3,995,707 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995587 120 + 27905053 AUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGUGGGCUGUUAAUGUUAAUCCCAAGUUAUAUGUGAACGUAUGUAAUGUAUUACACAUAUACGAGUAUAU .(((((.(((((((........))))))))))))(((..((((((.((.(((....))).)).))))))..))).((.(((((((...((((.....))))..)))))))))........ ( -35.10) >DroSec_CAF1 1382 94 + 1 AUUGGCAUGACGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGUAUGCUGUUAAUGUUAAUCCCAAGUUAUA--------------------------UAUACGAGUAUAU .(((((.((.((((........)))).)))))))(((..((((((.((.(((....))).)).))))))..)))....(((--------------------------(((....)))))) ( -21.10) >DroSim_CAF1 1381 94 + 1 AUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGCGGGCUGUUAAUGUUAAUCCCAAGUUAUA--------------------------UAUGCGAGUAUAU .(((((.(((((((........))))))))))))(((..((((((.((.(((....))).)).))))))..)))....(((--------------------------(((....)))))) ( -28.70) >DroYak_CAF1 832 108 + 1 AUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGCGGGCUGUUAAUGUUAAUCC-AAGCUAUAUGGCAACUUAUAU--UG---------UAUUCUCGUAUAU .(((((.(((((((........))))))))))))(((.(((((((.((.(((....))).)).)))))))))-)...((((((((((......)--))---------).....)))))). ( -28.60) >consensus AUUGGCAUGGCGAAACUUUCAAUUUGCCAGCCGAUGGUGUUGGCGAUAUCAGCGGGCUGUUAAUGUUAAUCCCAAGUUAUA__________________________UAUACGAGUAUAU .(((((.(((((((........))))))))))))(((..((((((.((.(((....))).)).))))))..))).............................................. (-23.98 = -24.48 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:41 2006