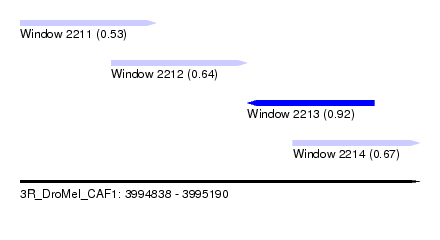
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,994,838 – 3,995,190 |
| Length | 352 |
| Max. P | 0.921867 |

| Location | 3,994,838 – 3,994,958 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -16.39 |
| Energy contribution | -17.57 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3994838 120 + 27905053 CAUUGUACGCCAAGAUGGAUUAGGCCAAAUCAAGACGAGAGUCGAAAGGAACAGCGUGAGGGAAACUUUAAAUAACGAUGGGUUGGAAGAAUAGGCAAUGCUCUAACAUCAAAAAAAAAA ((((((((((.....(((......)))......(((....)))..........))))((((....)))).....)))))).((((((..............))))))............. ( -25.24) >DroSec_CAF1 636 117 + 1 CAUUGCACGCCAAGAUGGAUUAGACCAAAUCAAGACGAGAGUCGAAAGGAACAGCGUGAGGGAAACUUUAAAUAACGAUGGGUCGGAAGAAUAGGCAAUGCUCUGAAAUCAAA---AAAA ((((((...((.....))....((((..(((..(((....))).............(((((....)))))......))).))))..........)))))).............---.... ( -26.50) >DroSim_CAF1 635 117 + 1 CAUUGCACGCCAAGAUGGAUUAGGCCAAAUCAAGACGAGAGUCGAAAGGAACAGCGUGAGGGAAACUCUAAAUAACGAUGGGUUGGAAGAAUAGGCAAUGCUCUAACAUCAAA---AAAA ((((((((((.....(((......)))......(((....)))..........)))))(((....).))......))))).((((((..............))))))......---.... ( -24.24) >DroYak_CAF1 73 103 + 1 CAUUGCACGCCGAGAUGGAUUAGGCCAAAUCAAGACGAGAGUCGAGAGG-------------UAACUUUAAAUAGCAAGGGGUUGGAGGAAUGGGGAAUGCUCUAACAUCAAA----AAA ..((((.....((..(((......)))..))..(((....))).((((.-------------...)))).....))))(..(((((((.(........).)))))))..)...----... ( -23.50) >consensus CAUUGCACGCCAAGAUGGAUUAGGCCAAAUCAAGACGAGAGUCGAAAGGAACAGCGUGAGGGAAACUUUAAAUAACGAUGGGUUGGAAGAAUAGGCAAUGCUCUAACAUCAAA___AAAA ((((((...((.....))....((((.......(((....))).............(((((....)))))..........))))..........)))))).................... (-16.39 = -17.57 + 1.19)


| Location | 3,994,918 – 3,995,038 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.642069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3994918 120 + 27905053 GGUUGGAAGAAUAGGCAAUGCUCUAACAUCAAAAAAAAAAUCAUAACUAAGUGAUUUUACUAUUGCAUUUUCCGCAUUGCAAUCAAAAAUCAAGAGCAGCAUAAUUAGUUGCUUAGACCC ((((..........(((((((........(((....((((((((......))))))))....)))........))))))).............(((((((.......))))))).)))). ( -27.39) >DroSec_CAF1 716 117 + 1 GGUCGGAAGAAUAGGCAAUGCUCUGAAAUCAAA---AAAAUCAUAACUAAGUGAUUUGAUCAUUGCAUUUUCCGUAUUGCAAUCAAAGAUCAAGAUCAGCAUAAUUAGUUGCUUAGACCC ((((((((((....((((((.((((....))..---.(((((((......))))))))).)))))).))))))..............(((....)))((((........))))..)))). ( -28.10) >DroSim_CAF1 715 117 + 1 GGUUGGAAGAAUAGGCAAUGCUCUAACAUCAAA---AAAAUCAUAACUAAGUGAUUUGAUCAUUGCAUUUUCCGUAUUGCAAUCAAAGAUCAAGAUCAGCAUAAUUAGUUGCUUAGACCC ((((((((((....((((((.((..........---.(((((((......))))))))).)))))).))))))..............(((....)))((((........))))..)))). ( -23.70) >DroYak_CAF1 140 110 + 1 GGUUGGAGGAAUGGGGAAUGCUCUAACAUCAAA----AAAUCAUAACUAAGUGAUUUGAUCAUGCCAUUUUCUGAAUUAUAAUCAAGCAUUAAGAUCAGCAUAAUUAGUAGCUU------ ((((.(..(((((((((....)))..(((....----(((((((......)))))))....)))))))))..).))))......(((((((((...........))))).))))------ ( -17.80) >consensus GGUUGGAAGAAUAGGCAAUGCUCUAACAUCAAA___AAAAUCAUAACUAAGUGAUUUGAUCAUUGCAUUUUCCGUAUUGCAAUCAAAGAUCAAGAUCAGCAUAAUUAGUUGCUUAGACCC ((((((((((....((((((.((..............(((((((......))))))))).)))))).))))))........................((((........))))..)))). (-17.12 = -17.80 + 0.69)


| Location | 3,995,038 – 3,995,150 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -21.50 |
| Energy contribution | -23.12 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995038 112 - 27905053 --------CCAACGAUCGGGUUUGGUUAUGUUUGCCAUCCACCGAUCAAUAAAUACCCUGCCUUGAGGUGGCAGCUACCACUUUGCGGGAUAAUUUCAGACAUCGAGUUGCAUAAUAAUU --------.(((((((((((..((((.......))))..).)))))).........(((((...(((((((......)))))))))))).................)))).......... ( -34.50) >DroSec_CAF1 833 112 - 1 --------CCAUCGAUCGGGUUUGGUUAUGUUCGCCAUUCACCGAUCACUAAAUACCCUCCCUUGAGGUGGCAGCUACCACUUUGCGGGAUAAUUCCAGACAUAGAGUUGCAUAAUAACU --------.....(((((((..((((.......))))..).))))))...........((((.((((((((......)))))))).))))((((((........)))))).......... ( -32.30) >DroSim_CAF1 832 112 - 1 --------CCAUCGAUCGGGUUUGGUUAUGUUUGCCAUCCACCGAUCACUAAAUACCCUGCCUUGAGGUGGCAGCUACCACUUUGCGGGAUAAUUCCAGACAUAGAGUUGCAUAAUAACU --------.....(((((((..((((.......))))..).)))))).........(((((...(((((((......)))))))))))).((((((........)))))).......... ( -33.30) >DroYak_CAF1 250 102 - 1 ACAACGAACCAUCGAUCGGGUUCAUGUAUGCUUGCCUUCUACCGAUCACUGAAUACCCUUUCUUGAGGUGGCAGUUACCAUUUUCUGGCAUAAUUUCAGAAA------------------ ..(((...(((((((((((......(((....)))......))))))...(((......)))....)))))..))).....(((((((.......)))))))------------------ ( -23.10) >consensus ________CCAUCGAUCGGGUUUGGUUAUGUUUGCCAUCCACCGAUCACUAAAUACCCUGCCUUGAGGUGGCAGCUACCACUUUGCGGGAUAAUUCCAGACAUAGAGUUGCAUAAUAACU .............(((((((..((((.......))))..).)))))).........(((((...(((((((......))))))))))))............................... (-21.50 = -23.12 + 1.62)



| Location | 3,995,078 – 3,995,190 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -28.77 |
| Energy contribution | -29.40 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3995078 112 + 27905053 UGGUAGCUGCCACCUCAAGGCAGGGUAUUUAUUGAUCGGUGGAUGGCAAACAUAACCAAACCCGAUCGUUGG--------AUCGCUGCUUUGGCCGGAUCGCUGGCUGCGAUAAUAUCCA ..((((((((..(((((((((((.(.(((((.(((((((.(..(((.........)))..)))))))).)))--------))).)))))))))..))...)).))))))........... ( -41.30) >DroSec_CAF1 873 112 + 1 UGGUAGCUGCCACCUCAAGGGAGGGUAUUUAGUGAUCGGUGAAUGGCGAACAUAACCAAACCCGAUCGAUGG--------AUCGCUGCUUUGGCCGGAUCGCUGGCUGCGAUAAUAUCCA .((((((..(((((((....))))........(((((((.(..(((.........)))..)))))))).)))--------...))))))..((((((....))))))............. ( -35.60) >DroSim_CAF1 872 112 + 1 UGGUAGCUGCCACCUCAAGGCAGGGUAUUUAGUGAUCGGUGGAUGGCAAACAUAACCAAACCCGAUCGAUGG--------AUCGCUGCUUUGGCCGGAUCGCUGGCUGCGAUAAUAUCCA ..((((((((..(((((((((((.(.(((((.(((((((.(..(((.........)))..)))))))).)))--------))).)))))))))..))...)).))))))........... ( -41.30) >DroYak_CAF1 272 120 + 1 UGGUAACUGCCACCUCAAGAAAGGGUAUUCAGUGAUCGGUAGAAGGCAAGCAUACAUGAACCCGAUCGAUGGUUCGUUGUAUCGCUGCUUUGGUCGGAUCGCUGUCUGCGAUAAUAUCCA ((((....))))(((......)))(((..((((((((.(...((((((.(((((((((((((........)))))).))))).))))))))...).))))))))..)))........... ( -41.40) >consensus UGGUAGCUGCCACCUCAAGGCAGGGUAUUUAGUGAUCGGUGGAUGGCAAACAUAACCAAACCCGAUCGAUGG________AUCGCUGCUUUGGCCGGAUCGCUGGCUGCGAUAAUAUCCA ((((....))))..........(((((((((.(((((((.(..(((.........)))..)))))))).)).........(((((......((((((....)))))))))))))))))). (-28.77 = -29.40 + 0.63)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:34 2006