
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,983,246 – 3,983,345 |
| Length | 99 |
| Max. P | 0.778897 |

| Location | 3,983,246 – 3,983,345 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -20.22 |
| Energy contribution | -21.78 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
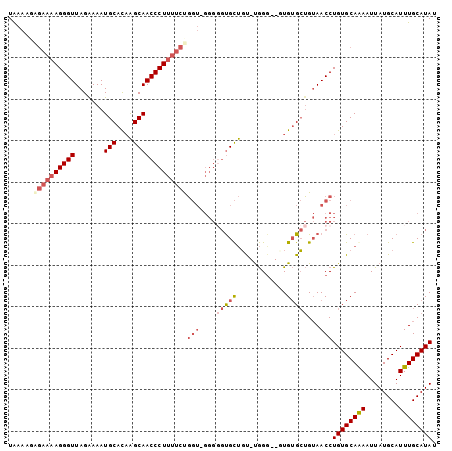
>3R_DroMel_CAF1 3983246 99 + 27905053 UAAAAGAGAAAAGGGUUAGAAAAUGCACAAGCAACCCUUUUCUGGU-GGGGGUGCUGU-CGGC--UUGUGCUGUAACCUGUGCAGAAUUAUGCAUUUGCAUAU ......((((((((((.......(((....))))))))))))).((-(.((((.....-((((--....))))..)))).))).....(((((....))))). ( -31.51) >DroSec_CAF1 29647 99 + 1 UAAAAGAGAAAAGGGUUAGAAAAUGCACAAGCAACCCUUUUCUGGU-GGGGGUGCUGU-UGGG--UUGUGCUGUAACCUGUGCAGAAUUAUGCAUUUGCAUAU ......((((((((((.......(((....))))))))))))).((-(.((((((...-....--(((..(........)..)))......)))))).))).. ( -29.33) >DroSim_CAF1 31336 100 + 1 UAAAAGAGAAAAGGGUUAGGAAAUGCACAAGCAACCCUUUUCUGGUUGGGGGUGCUGU-UGGG--UUGUGCUGUAACCUGUGCAGAAUUAUGCAUUUGCAUAU ......((((((((((.......(((....)))))))))))))(((((.(.(..(...-....--..)..)).)))))((((((((........)))))))). ( -29.11) >DroEre_CAF1 30460 99 + 1 UAAAAACGAAAAGGGUUAGAAAAUGCAUAAGCAACCCUUUUCUGGU-GGGGGCGCUGU-UGGG--GUGUGCUGUAACCUGUGCAAAAUUAUGCAUUUGCAUAU .......(((((((((.......(((....)))))))))))).(((-.(.(((((...-....--..))))).).)))((((((((........)))))))). ( -30.41) >DroYak_CAF1 31760 101 + 1 UAAAAUGGAAAAGGGUUAGAAAAUGCAUAAGCAACCCUUUUCUGGU-GGG-GUGCUGUGUGGGGUGUGUGCUGUAACCUGUGCAAAAUUAUGCAUUUGCAUAU ......((((((((((.......(((....)))))))))))))(((-(.(-(..(............)..)).).)))((((((((........)))))))). ( -27.61) >DroPer_CAF1 33072 88 + 1 UGGA----AAAAGGGUUAGAAAAUGCACUGGCAACCCUCAACCC--------UGUCCC-UAGG--GUGGGGGGAAACCUGUGCAAAAUUAUGCAUUUGCAUAU ....----...............(((((.((...(((((.((((--------((....-))))--)).)))))...)).)))))....(((((....))))). ( -31.40) >consensus UAAAAGAGAAAAGGGUUAGAAAAUGCACAAGCAACCCUUUUCUGGU_GGGGGUGCUGU_UGGG__GUGUGCUGUAACCUGUGCAAAAUUAUGCAUUUGCAUAU ......((((((((((.......(((....)))))))))))))(((....(((((............)))))...)))((((((((........)))))))). (-20.22 = -21.78 + 1.56)


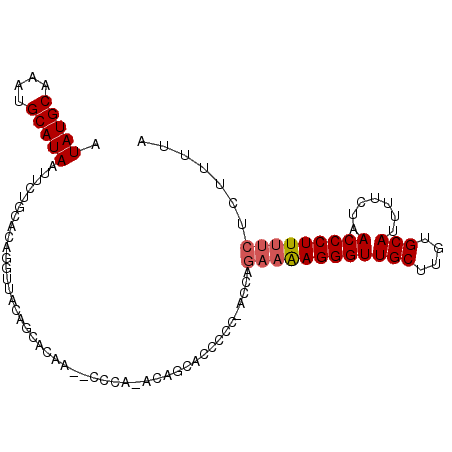
| Location | 3,983,246 – 3,983,345 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
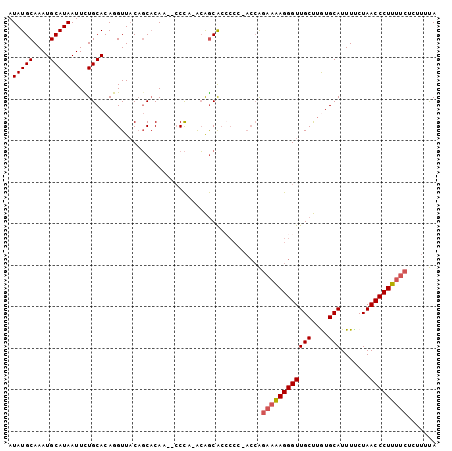
>3R_DroMel_CAF1 3983246 99 - 27905053 AUAUGCAAAUGCAUAAUUCUGCACAGGUUACAGCACAA--GCCG-ACAGCACCCCC-ACCAGAAAAGGGUUGCUUGUGCAUUUUCUAACCCUUUUCUCUUUUA .(((((....)))))....(((...((((........)--))).-...))).....-...(((((((((((((....))).......))))))))))...... ( -24.31) >DroSec_CAF1 29647 99 - 1 AUAUGCAAAUGCAUAAUUCUGCACAGGUUACAGCACAA--CCCA-ACAGCACCCCC-ACCAGAAAAGGGUUGCUUGUGCAUUUUCUAACCCUUUUCUCUUUUA .(((((....)))))....(((...((((.......))--))..-...))).....-...(((((((((((((....))).......))))))))))...... ( -22.11) >DroSim_CAF1 31336 100 - 1 AUAUGCAAAUGCAUAAUUCUGCACAGGUUACAGCACAA--CCCA-ACAGCACCCCCAACCAGAAAAGGGUUGCUUGUGCAUUUCCUAACCCUUUUCUCUUUUA .(((((....)))))....(((...((((.......))--))..-...))).........(((((((((((((....))).......))))))))))...... ( -22.11) >DroEre_CAF1 30460 99 - 1 AUAUGCAAAUGCAUAAUUUUGCACAGGUUACAGCACAC--CCCA-ACAGCGCCCCC-ACCAGAAAAGGGUUGCUUAUGCAUUUUCUAACCCUUUUCGUUUUUA ...((((((........))))))..(((....((.(..--....-...).))....-))).((((((((((((....))).......)))))))))....... ( -22.71) >DroYak_CAF1 31760 101 - 1 AUAUGCAAAUGCAUAAUUUUGCACAGGUUACAGCACACACCCCACACAGCAC-CCC-ACCAGAAAAGGGUUGCUUAUGCAUUUUCUAACCCUUUUCCAUUUUA ...((((((........))))))..(((....((..............))..-...-))).((((((((((((....))).......)))))))))....... ( -20.35) >DroPer_CAF1 33072 88 - 1 AUAUGCAAAUGCAUAAUUUUGCACAGGUUUCCCCCCAC--CCUA-GGGACA--------GGGUUGAGGGUUGCCAGUGCAUUUUCUAACCCUUUU----UCCA .(((((....)))))....(((((.(((..(((.(.((--(((.-.....)--------)))).).)))..))).)))))...............----.... ( -30.20) >consensus AUAUGCAAAUGCAUAAUUCUGCACAGGUUACAGCACAA__CCCA_ACAGCACCCCC_ACCAGAAAAGGGUUGCUUGUGCAUUUUCUAACCCUUUUCUCUUUUA .(((((....)))))..............................................((((((((((((....))).......)))))))))....... (-16.92 = -17.28 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:26 2006