
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,961,232 – 3,961,323 |
| Length | 91 |
| Max. P | 0.973557 |

| Location | 3,961,232 – 3,961,323 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -12.39 |
| Energy contribution | -13.44 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
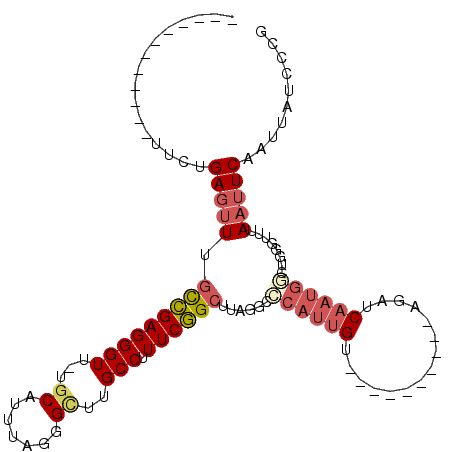
>3R_DroMel_CAF1 3961232 91 + 27905053 CGGGAUAAUUGAAUUAAACCCAGCCAUUGAUCU----------ACAAUGGAACUAAGCCGAAAGGCAAGCCCUAAACGCAAAACCCUCGGCAAACUCAGAA------------ .(((..............)))..((((((....----------.))))))......(((....)))..(((.................)))..........------------ ( -19.77) >DroSec_CAF1 7903 91 + 1 CGGGAUAAUUGAAUUAAACCCAGACAUUGAACU----------ACAAUGGAUCUAAGCCGAAAGGCAAGCCCUAAAUGAAAAACCCUCGGCAAACUCAGAA------------ .(((..............)))...(((((....----------.)))))..(((..(((....)))..(((.................)))......))).------------ ( -17.97) >DroSim_CAF1 9340 91 + 1 CGGGAUAAUUGAAUUAAACCCAGGCAUUGGACU----------ACAAUGCAACUAAGCCGAAAGGCAAGCCCUAAACGCAAAACCCUCGGCAAACUCAGAA------------ .(((..............)))..((((((....----------.))))))......(((....)))..(((.................)))..........------------ ( -21.57) >DroEre_CAF1 7904 97 + 1 CGGGAUAAUUGAAUUAAACCCAGCCAUUGAUCUACAAUGAUCUACAAUGGGCCUAAGCCGAAAGGCAAGCCCUAAAUGCA-AACCCUCGACAAACUCC--------------- .(((.........(((..((((......((((......)))).....))))..)))(((....)))..............-..)))............--------------- ( -23.60) >DroYak_CAF1 8181 102 + 1 CGGCAUAAUUGAAUUAAACCCAGCCAUUGAUCU----------ACAAUGGGCCUAAGCCGAAAGGCAAGCCCUAAAUGCA-AACCCUCGACAAACUCAGAAAAACCGAGAAGG .(((...................((((((....----------.))))))......(((....)))..))).........-....((((................)))).... ( -22.59) >DroPer_CAF1 10089 72 + 1 UGGCAUCAUC--AGUAA--------AUUGAAUU----------ACCGGGGGGCUAAGCUGAAAGGCAAACCCAAAAUGCA-AACCCUCAAAAA-------------------- ..((((..((--.((((--------......))----------)).))(((.....(((....)))...)))...)))).-............-------------------- ( -13.40) >consensus CGGGAUAAUUGAAUUAAACCCAGCCAUUGAACU__________ACAAUGGAACUAAGCCGAAAGGCAAGCCCUAAAUGCA_AACCCUCGACAAACUCAGAA____________ .(((...................((((((...............))))))......(((....))).................)))........................... (-12.39 = -13.44 + 1.06)



| Location | 3,961,232 – 3,961,323 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.63 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3961232 91 - 27905053 ------------UUCUGAGUUUGCCGAGGGUUUUGCGUUUAGGGCUUGCCUUUCGGCUUAGUUCCAUUGU----------AGAUCAAUGGCUGGGUUUAAUUCAAUUAUCCCG ------------...((((((.(((((((((...((.......))..))).))))))((((..((((((.----------....))))))))))....))))))......... ( -24.50) >DroSec_CAF1 7903 91 - 1 ------------UUCUGAGUUUGCCGAGGGUUUUUCAUUUAGGGCUUGCCUUUCGGCUUAGAUCCAUUGU----------AGUUCAAUGUCUGGGUUUAAUUCAAUUAUCCCG ------------...(((((..(((((((((..(((.....)))...))).)))))).(((((((((((.----------....)))....)))))))))))))......... ( -20.80) >DroSim_CAF1 9340 91 - 1 ------------UUCUGAGUUUGCCGAGGGUUUUGCGUUUAGGGCUUGCCUUUCGGCUUAGUUGCAUUGU----------AGUCCAAUGCCUGGGUUUAAUUCAAUUAUCCCG ------------....((.(..(((((((((...((.......))..))).))))))..).))((((((.----------....))))))..(((..((((...)))).))). ( -24.70) >DroEre_CAF1 7904 97 - 1 ---------------GGAGUUUGUCGAGGGUU-UGCAUUUAGGGCUUGCCUUUCGGCUUAGGCCCAUUGUAGAUCAUUGUAGAUCAAUGGCUGGGUUUAAUUCAAUUAUCCCG ---------------(((....(((((((((.-.((.......))..))).))))))((((((((((..(.((((......)))).)..).)))))))))........))).. ( -31.50) >DroYak_CAF1 8181 102 - 1 CCUUCUCGGUUUUUCUGAGUUUGUCGAGGGUU-UGCAUUUAGGGCUUGCCUUUCGGCUUAGGCCCAUUGU----------AGAUCAAUGGCUGGGUUUAAUUCAAUUAUGCCG ....(((((.....)))))...((((..((((-((((....(((((((((....)))..))))))..)))----------)))))..))))..(((.((((...)))).))). ( -32.40) >DroPer_CAF1 10089 72 - 1 --------------------UUUUUGAGGGUU-UGCAUUUUGGGUUUGCCUUUCAGCUUAGCCCCCCGGU----------AAUUCAAU--------UUACU--GAUGAUGCCA --------------------........(((.-..(((...(((((.((......))..)))))..((((----------((......--------)))))--))))..))). ( -17.40) >consensus ____________UUCUGAGUUUGCCGAGGGUU_UGCAUUUAGGGCUUGCCUUUCGGCUUAGGCCCAUUGU__________AGAUCAAUGGCUGGGUUUAAUUCAAUUAUCCCG ................(((((.(((((((((...((.......))..))).))))))......((((((...............))))))........))))).......... (-13.68 = -14.63 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:12 2006