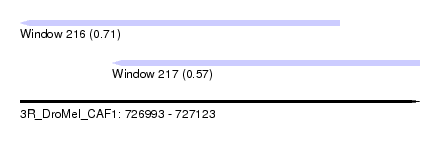
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 726,993 – 727,123 |
| Length | 130 |
| Max. P | 0.705839 |

| Location | 726,993 – 727,097 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.85 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -26.45 |
| Energy contribution | -25.78 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
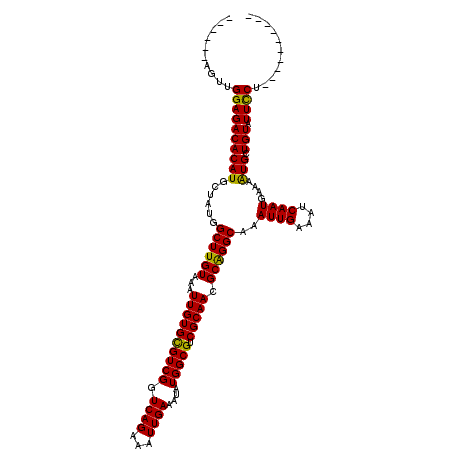
>3R_DroMel_CAF1 726993 104 - 27905053 ------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAAGGCAGGCAAAUUGAAAUCAAUGAAAAUGCUGUAUUUCU---------- ------....(((((.(((.((....((((((...((((((((((.((((...))))....))))).))))).))))))..((((....))))......))))).)))))---------- ( -29.30) >DroPse_CAF1 143922 120 - 1 GGGGGCCGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCAAAUUGAAAUCAAUGAAAAUGCUGUAUUCCUCUGACUAAAG .......((..(((..(((.((....((((((...((((((((((.((((...))))....))))).))))).))))))..((((....))))......)))))....)))..))..... ( -30.70) >DroSec_CAF1 138909 104 - 1 ------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAAGGCGGGCAAAUUGAAAUCAAUGAAAGUGCUGUAUUUCU---------- ------....(((((.(((.((....((((((...((((((((((.((((...))))....))))).))))).))))))..((((....))))......))))).)))))---------- ( -28.50) >DroEre_CAF1 136834 104 - 1 ------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGUGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCAAAUUGAAAUCAAUGAAAAUGCUGUAUUUCU---------- ------..((((.(((((((((....)).........)))))))..)))).....(((((((((((((........)))..((((....))))......)))))))))).---------- ( -25.40) >DroAna_CAF1 138582 104 - 1 ------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCAAAUUGAAAUCAAUGAAAAUGCUGUAUUCCU---------- ------....((((((((((......((((((...((((((((((.((((...))))....))))).))))).))))))..((((....))))....))).))).)))).---------- ( -26.90) >DroPer_CAF1 144139 120 - 1 GGGGGCCAUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCAAAUUGAAAUCAAUGAAAAUGCUGUAUUCCUCUGACUAAAG (((((.....(....)(((.((....((((((...((((((((((.((((...))))....))))).))))).))))))..((((....))))......)))))..)))))......... ( -29.90) >consensus ______AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCAAAUUGAAAUCAAUGAAAAUGCUGUAUUCCU__________ ..........((((((((((......((((((...((((((((((.((((...))))....))))).))))).))))))..((((....))))....))).))).))))........... (-26.45 = -25.78 + -0.67)

| Location | 727,023 – 727,123 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -24.32 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
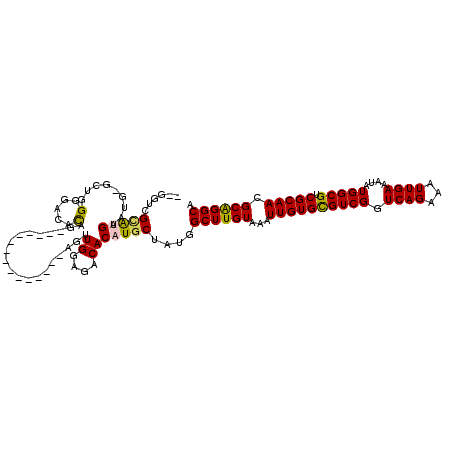
>3R_DroMel_CAF1 727023 100 - 27905053 --GGUCGCAUGAAUG-GCUGGGGCCCCAG---------------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAAGGCAGGCA --....(((((....-.((((....))))---------------...((....)))))))....((((((...((((((((((.((((...))))....))))).))))).)))))). ( -36.60) >DroPse_CAF1 143962 118 - 1 AUGGGGGUACGAGUGAGCAUGGGGUACGGACGGGGGGUGGGGGCCGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCA .((.(..(((((((.((((((..((.(.(((((..........))))).)..)).))))))...)))))))..((((((((((.((((...))))....))))).)))))).)).... ( -36.20) >DroSim_CAF1 141132 100 - 1 --GGUCGCAUGAAUG-GCUGGGGACACAG---------------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAAGGCGGGCA --....(((((....-.(((......)))---------------...((....)))))))....((((((...((((((((((.((((...))))....))))).))))).)))))). ( -32.20) >DroEre_CAF1 136864 97 - 1 --GGUCGCAUGAAUG-GCUGGG---CCAG---------------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGUGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCA --....(((((..((-((...)---))).---------------...((....)))))))....((((((...((((((((((.((((...))))....))))).))))).)))))). ( -30.10) >DroAna_CAF1 138612 98 - 1 --GGGUG-GUGGAUG-GA-GGGCAGAUGG---------------AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCA --.....-.......-..-.((((..((.---------------.........))..))))...((((((...((((((((((.((((...))))....))))).))))).)))))). ( -23.90) >DroPer_CAF1 144179 117 - 1 AUGG-GGUACGAGUGAGCAUGGGGUACGGACGGGGGGUGGGGGCCAUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCA .((.-(.(((((((.((((((.((..(..((.....))..)..))..((....))))))))...)))))))..((((((((((.((((...))))....))))).)))))).)).... ( -35.00) >consensus __GGUCGCAUGAAUG_GCUGGGGACACAG_______________AGUUGGAGACACAUGCUAUGGCUUGUAAAUUGUGCGUCGGUCAGAAAUUGAAAUAUGGCGUCGCAACGCAGGCA ......(((((.........(.....)....................((....)))))))....((((((...((((((((((.((((...))))....))))).))))).)))))). (-24.32 = -24.10 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:02 2006