
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,948,705 – 3,948,835 |
| Length | 130 |
| Max. P | 0.995902 |

| Location | 3,948,705 – 3,948,816 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -24.76 |
| Energy contribution | -25.82 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3948705 111 + 27905053 UCGAAAUGACUACGGCGGAGAAGGACGAGGCUAUCGCGCUAAACGCAAUGAUUAUGGCGGUGGACGUCGCGACAAUCGGGAUAA---------UUACGGAGCAGGGCAUCGUGAUAGUCG (((..(((.((((((((..((.((......)).)).))))...(((..........))))))).)))..))).......(((.(---------(((((..((...))..)))))).))). ( -27.70) >DroSim_CAF1 17209 111 + 1 GCGAAAUGACAACGGCGGAGAAGGACGAGGCUAUCGCGGUAAACGCGAUGAUUACGGCGGUGGACGUCGCGACUAUCGUGAUAA---------UUACGGAGCAGGCUGUCGUGAUAGUCG .......(((.(((((((.......((..(.((((((((((..(((((((.((((....)))).)))))))..)))))))))).---------)..)).......)))))))....))). ( -40.54) >DroEre_CAF1 17161 120 + 1 UCGUGAUGACUACGGCGGAGGAAGACGAGAUUAUCGCGGUAAACGCGAUGGUUACGGCGGAGGACGCCGCGACGAUCGGGAUAACCAAGAUAAUUACGGCGCAGGGCGUCAUGAUAGUCG ((((((((.((.((.((..(.....)..(((((((..(((...(.((((.((..(((((.....)))))..)).)))).)...)))..))))))).)).)).))..))))))))...... ( -46.80) >DroYak_CAF1 17299 103 + 1 -----------------GAGGAGGACGAGAUUAUCGCGGUAAACGCGAUGAUUAUGGCGGAGGACGUCGCGCCCAUCGGGAAAAUAGAGAUGUCUACGGCGCAGUGCGACGUGAUAGUCG -----------------......(((..((((((((((.....))))))))))((.(((..(.((...(((((((((...........)))).....))))).)).)..))).)).))). ( -37.10) >consensus UCGAAAUGACUACGGCGGAGAAGGACGAGACUAUCGCGGUAAACGCGAUGAUUACGGCGGAGGACGUCGCGACAAUCGGGAUAA_________UUACGGAGCAGGGCGUCGUGAUAGUCG ............................(((((((((......(.(((((.(..(((((.....)))))..).))))).).................(((((...)))))))))))))). (-24.76 = -25.82 + 1.06)



| Location | 3,948,745 – 3,948,835 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.61 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -16.35 |
| Energy contribution | -17.63 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3948745 90 + 27905053 AAACGCAAUGAUUAUGGCGGUGGACGUCGCGACAAUCGGGAUAA---------UUACGGAGCAGGGCAUCGUGAUAGUCGCGACAGUGCCGGGGGA------GGA ...(((..........))).(((((((((((((.((((.(((..---------((........))..))).)))).)))))))).)).))).....------... ( -26.70) >DroSec_CAF1 16708 81 + 1 ---------------UGCGGUGGACGUCGCGACUAUCGUGAUAA---------UUACGGAGCGGGCUGUCGUGAUAGUCGCGACAGCGGUAGUGGAGGAGGAGGA ---------------(((.((....((((((((((((((((((.---------...((...))...)))))))))))))))))).)).))).............. ( -29.80) >DroSim_CAF1 17249 93 + 1 AAACGCGAUGAUUACGGCGGUGGACGUCGCGACUAUCGUGAUAA---------UUACGGAGCAGGCUGUCGUGAUAGUCGCGACAGCGGUGGUGGAGGA---GGA ...(((((((.((((....)))).)))))))((((((((((...---------)))).......(((((((((.....)))))))))))))))......---... ( -36.80) >DroEre_CAF1 17201 98 + 1 AAACGCGAUGGUUACGGCGGAGGACGCCGCGACGAUCGGGAUAACCAAGAUAAUUACGGCGCAGGGCGUCAUGAUAGUCGCGACGGU------GGAGGAGGAGG- ...((((((.((..(((((.....)))))..)).)))((.....))..(((.((((.(((((...))))).)))).)))))).....------...........- ( -34.80) >DroYak_CAF1 17322 99 + 1 AAACGCGAUGAUUAUGGCGGAGGACGUCGCGCCCAUCGGGAAAAUAGAGAUGUCUACGGCGCAGUGCGACGUGAUAGUCGCGACGGU------GGAGGAGGAGGA ...((((.(((((((.(((..(.((...(((((((((...........)))).....))))).)).)..))).))))))))).))..------............ ( -31.40) >consensus AAACGCGAUGAUUACGGCGGUGGACGUCGCGACUAUCGGGAUAA_________UUACGGAGCAGGGCGUCGUGAUAGUCGCGACAGUG___G_GGAGGAGGAGGA ...(((..........)))......((((((((.((((.(((.........................))).)))).))))))))..................... (-16.35 = -17.63 + 1.28)



| Location | 3,948,745 – 3,948,835 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.61 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3948745 90 - 27905053 UCC------UCCCCCGGCACUGUCGCGACUAUCACGAUGCCCUGCUCCGUAA---------UUAUCCCGAUUGUCGCGACGUCCACCGCCAUAAUCAUUGCGUUU ...------......((.((.((((((((.(((..((((...(((...))).---------.))))..))).))))))))))))..(((.((....)).)))... ( -22.70) >DroSec_CAF1 16708 81 - 1 UCCUCCUCCUCCACUACCGCUGUCGCGACUAUCACGACAGCCCGCUCCGUAA---------UUAUCACGAUAGUCGCGACGUCCACCGCA--------------- ..................(((((((((((((((.((......))....((..---------.....)))))))))))))))......)).--------------- ( -21.60) >DroSim_CAF1 17249 93 - 1 UCC---UCCUCCACCACCGCUGUCGCGACUAUCACGACAGCCUGCUCCGUAA---------UUAUCACGAUAGUCGCGACGUCCACCGCCGUAAUCAUCGCGUUU ...---...............(((((((((((((((..((....)).)))..---------.......))))))))))))......(((.((....)).)))... ( -22.90) >DroEre_CAF1 17201 98 - 1 -CCUCCUCCUCC------ACCGUCGCGACUAUCAUGACGCCCUGCGCCGUAAUUAUCUUGGUUAUCCCGAUCGUCGCGGCGUCCUCCGCCGUAACCAUCGCGUUU -...........------..(((((((((.((((((.(((...))).))).........((.....))))).))))))))).....(((.((....)).)))... ( -25.00) >DroYak_CAF1 17322 99 - 1 UCCUCCUCCUCC------ACCGUCGCGACUAUCACGUCGCACUGCGCCGUAGACAUCUCUAUUUUCCCGAUGGGCGCGACGUCCUCCGCCAUAAUCAUCGCGUUU ............------..(((((((((......)))))...((((((((((....)))))..........))))))))).....(((.((....)).)))... ( -25.70) >consensus UCCUCCUCCUCC_C___CACUGUCGCGACUAUCACGACGCCCUGCUCCGUAA_________UUAUCCCGAUAGUCGCGACGUCCACCGCCAUAAUCAUCGCGUUU ....................(((((((((((((...................................)))))))))))))........................ (-17.65 = -17.77 + 0.12)


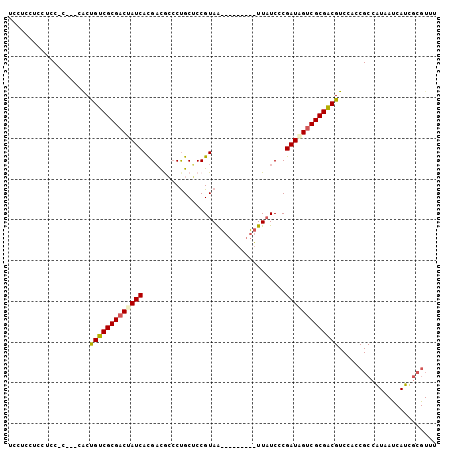
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:07 2006