
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,904,866 – 3,904,965 |
| Length | 99 |
| Max. P | 0.635385 |

| Location | 3,904,866 – 3,904,965 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.02 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
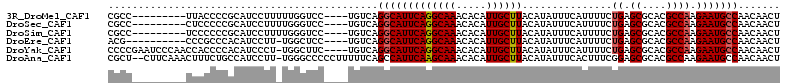
>3R_DroMel_CAF1 3904866 99 + 27905053 CGCC---------UUACCCCGCAUCCUUUUUGGUCC----UGUCAGGCAUUCAGGCAAACACAUUGCUUACAUAUUUCAUUUUCUGAGCGCACGCCAAGAAUGCCAACAACU ....---------.......(((.((.....))...----)))..(((((((((((((.....))))))...............((.((....)))).)))))))....... ( -20.00) >DroSec_CAF1 28066 99 + 1 CGCC---------CUCCCCCGCAUCCUUUUGGGUCC----UGUCAGGCAUUCAGGCAAACACAUUGCUUACAUAUUUCAUUUUCUGAGCGCACGCCAAGAAUGCCAACAACU .(((---------(................))))..----.....(((((((((((((.....))))))...............((.((....)))).)))))))....... ( -22.49) >DroSim_CAF1 28469 99 + 1 CGCC---------UCCCCCCGCAUCCUUUUGGGUCC----UGUCAGGCAUUCAGGCAAACACAUUGCUUACAUAUUUCAUUUUCUGAGCGCACGCCAAGAAUGCCAACAACU ....---------....((((........))))...----.....(((((((((((((.....))))))...............((.((....)))).)))))))....... ( -22.00) >DroEre_CAF1 28038 97 + 1 ACG----------CCCGCCCACAUCCUU-UGGCUCC----UGUCAGGCAUUCAGGCAAACACAUUGCUUACAUAUUUCAUUUUCUGAGCGCACGCCAAGAAUGCCAACAACU ...----------...............-((((...----.))))(((((((((((((.....))))))...............((.((....)))).)))))))....... ( -21.50) >DroYak_CAF1 29756 107 + 1 CCCCGAAUCCCAACCACCCCACAUCCCU-UGGCUUC----UGUCAGGCAUUCAGGCAAACACAUUGCUUACAUAUUUCAUUUUCUGAGCGCACGCCAAGAAUGCCAACAACU ............................-((((...----.))))(((((((((((((.....))))))...............((.((....)))).)))))))....... ( -21.50) >DroAna_CAF1 29117 109 + 1 CGCU--CUUCAAACUUUCUGCCAUCCUU-UGGGCCCCCUUUUUCAGCCAUUCAAGCAAACACAUUGCUUACAUAUUUCACUUUCGGAGCGCACGCCAAGAAUGCCAACAACU ((((--((...........(((......-..)))..................((((((.....))))))...............))))))...................... ( -16.00) >consensus CGCC_________CCCCCCCGCAUCCUU_UGGGUCC____UGUCAGGCAUUCAGGCAAACACAUUGCUUACAUAUUUCAUUUUCUGAGCGCACGCCAAGAAUGCCAACAACU .............................................(((((((((((((.....))))))...............((.((....)))).)))))))....... (-16.74 = -16.93 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:54 2006