
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,897,529 – 3,897,669 |
| Length | 140 |
| Max. P | 0.999910 |

| Location | 3,897,529 – 3,897,635 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 98.49 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -30.13 |
| Energy contribution | -30.33 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
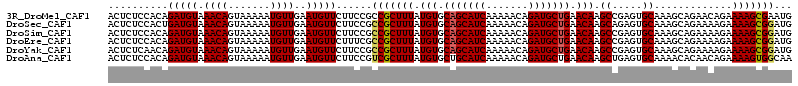
>3R_DroMel_CAF1 3897529 106 + 27905053 CUUCCGCUACCCCUUGUUCCAGAUCCCAUUCGCUUUUCUGUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACA ((((((((...........((((.............))))....(((((((.....)).(((.(((((((.......))))))).))).))))))))))))).... ( -33.42) >DroSec_CAF1 20734 106 + 1 CUUCCGCUACCCCUUGUUCCAGAUCCCAUCCGCUUUUCUUUUCUGCUUUGCACUCUGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACA ((((((((......(((..((((..................))))....)))....((((((.(((((((.......))))))).)))...))))))))))).... ( -31.47) >DroSim_CAF1 21177 106 + 1 CUUCCGCUACCCCUUGUUCCAGAUCCCAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACA ((((((((......(((..((((..................))))....)))....((((((.(((((((.......))))))).)))...))))))))))).... ( -31.47) >DroEre_CAF1 19572 106 + 1 CUUCCGCUACCCCUUGUUCCAGAUCCCAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGAAAGAACA ...................................((((((((((((((((.....)).(((.(((((((.......))))))).))).))))))).))))))).. ( -26.20) >DroYak_CAF1 22272 106 + 1 CUUCCGCUACCCCUUGUUCCAGAUCCCAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACA ((((((((......(((..((((..................))))....)))....((((((.(((((((.......))))))).)))...))))))))))).... ( -31.47) >consensus CUUCCGCUACCCCUUGUUCCAGAUCCCAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACA ((((((((......(((..((((..................))))....)))....((((((.(((((((.......))))))).)))...))))))))))).... (-30.13 = -30.33 + 0.20)



| Location | 3,897,529 – 3,897,635 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 98.49 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -31.34 |
| Energy contribution | -31.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3897529 106 - 27905053 UGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAACAGAAAAGCGAAUGGGAUCUGGAACAAGGGGUAGCGGAAG ....(((((((..(((..(((.(((((((.......)))))))......(((..(((....(......)....)))..)))........)))..)))..))))))) ( -32.80) >DroSec_CAF1 20734 106 - 1 UGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCAGAGUGCAAAGCAGAAAAGAAAAGCGGAUGGGAUCUGGAACAAGGGGUAGCGGAAG ....(((((((..(((..(((.(((((((.......)))))))......((((.(.((..((...........))...)).).))))..)))..)))..))))))) ( -34.30) >DroSim_CAF1 21177 106 - 1 UGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUGGGAUCUGGAACAAGGGGUAGCGGAAG ....(((((((..(((..(((.(((((((.......)))))))......(((..(.(...((...........)).....).)..))).)))..)))..))))))) ( -32.40) >DroEre_CAF1 19572 106 - 1 UGUUCUUUCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUGGGAUCUGGAACAAGGGGUAGCGGAAG (((((((((((((((((.(((.(((((((.......))))))).))).((.....)).............))))))).))))....)))))).............. ( -29.50) >DroYak_CAF1 22272 106 - 1 UGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUGGGAUCUGGAACAAGGGGUAGCGGAAG ....(((((((..(((..(((.(((((((.......)))))))......(((..(.(...((...........)).....).)..))).)))..)))..))))))) ( -32.40) >consensus UGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUGGGAUCUGGAACAAGGGGUAGCGGAAG ....(((((((((((((.(((.(((((((.......))))))).))).((.....)).............))))))......((((........)))).))))))) (-31.34 = -31.18 + -0.16)

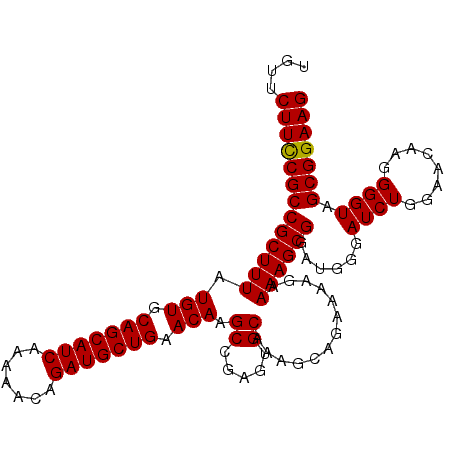
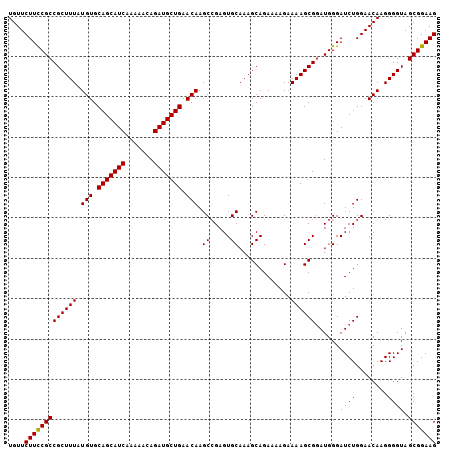
| Location | 3,897,555 – 3,897,669 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -31.19 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3897555 114 + 27905053 CAUUCGCUUUUCUGUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACAUUCAACAUUUUUACUGUUUACAUCUGUGGAGAGU .....(.(((((((..(((((((((.....)).(((.(((((((.......))))))).))).)))))))))))))).).............(((.(((((....))))).))) ( -32.20) >DroSec_CAF1 20760 114 + 1 CAUCCGCUUUUCUUUUCUGCUUUGCACUCUGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACAUUCAACAUUUUUACUGUUUACAUCAGUGGAGAGU .........((((((((((((((((.....)).(((.(((((((.......))))))).))).))))))).))))))).........(((((((((.......))))))))).. ( -33.50) >DroSim_CAF1 21203 114 + 1 CAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACAUUCAACAUUUUUACUGUUUACAUCUGUGGAGAGU ..(((((..((((((((((((((((.....)).(((.(((((((.......))))))).))).))))))).))))))).....((((.......)))).......))))).... ( -32.10) >DroEre_CAF1 19598 114 + 1 CAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGAAAGAACAUUCAACAUUUUUACUGUUUACAUCUGUGGAGAGU ..(((((..((((((((((((((((.....)).(((.(((((((.......))))))).))).))))))).))))))).....((((.......)))).......))))).... ( -32.50) >DroYak_CAF1 22298 114 + 1 CAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACAUUCAACAUUUUUACUGUUUACAUCUGUUGAGAGU .....((((((((((((((((((((.....)).(((.(((((((.......))))))).))).))))))).)))))))...((((((.................)))))))))) ( -30.23) >DroAna_CAF1 22436 114 + 1 UUGCCACUUUUCUGUUGUGUUUUGCACUCAGCUUGUUCAGCAUCUGUUUUUGAUGCAGCACAUAAAGCGACGGAAGAACAUUCAACAUUUUUACUGUUUACAUCUGUGGAGAGU .......((((((((((((.....)))...((((((.(.(((((.......))))).).)))...))))))))))))...............(((.(((((....))))).))) ( -26.60) >consensus CAUCCGCUUUUCUUUUCUGCUUUGCACUCGGCUUGUUCAGCAUCUGUUUUUGAUGCUGCACAUAAAGCGGCGGAAGAACAUUCAACAUUUUUACUGUUUACAUCUGUGGAGAGU .........((((((((((((((((.....)).(((.(((((((.......))))))).))).))))))).)))))))..............(((.(((((....))))).))) (-28.26 = -28.82 + 0.56)


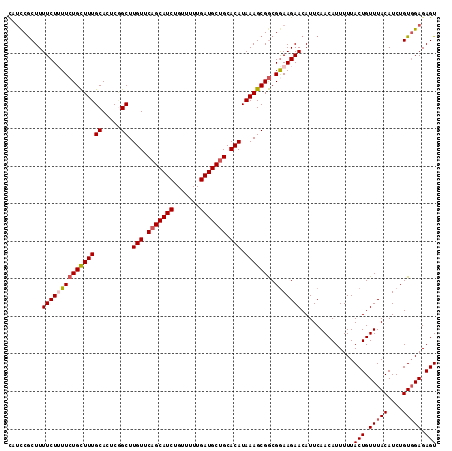
| Location | 3,897,555 – 3,897,669 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3897555 114 - 27905053 ACUCUCCACAGAUGUAAACAGUAAAAAUGUUGAAUGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAACAGAAAAGCGAAUG ...........................((((...((((((......(((((.(((.(((((((.......))))))).))).((.....)))))))))))))....)))).... ( -24.30) >DroSec_CAF1 20760 114 - 1 ACUCUCCACUGAUGUAAACAGUAAAAAUGUUGAAUGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCAGAGUGCAAAGCAGAAAAGAAAAGCGGAUG ....(((((((.......))))......(((.....(((((...(.(((((.(((.(((((((.......))))))).))).((.....))))))).)..))))).)))))).. ( -28.30) >DroSim_CAF1 21203 114 - 1 ACUCUCCACAGAUGUAAACAGUAAAAAUGUUGAAUGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUG ....(((.(.......((((.......)))).....(((((...(.(((((.(((.(((((((.......))))))).))).((.....))))))).)..)))))..).))).. ( -25.30) >DroEre_CAF1 19598 114 - 1 ACUCUCCACAGAUGUAAACAGUAAAAAUGUUGAAUGUUCUUUCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUG ....(((.(.......((((.......)))).....((((((..(.(((((.(((.(((((((.......))))))).))).((.....))))))).).))))))..).))).. ( -25.70) >DroYak_CAF1 22298 114 - 1 ACUCUCAACAGAUGUAAACAGUAAAAAUGUUGAAUGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUG ((..((((((....((.....))....))))))..)).......(((((((.(((.(((((((.......))))))).))).((.....)).............)))))))... ( -28.30) >DroAna_CAF1 22436 114 - 1 ACUCUCCACAGAUGUAAACAGUAAAAAUGUUGAAUGUUCUUCCGUCGCUUUAUGUGCUGCAUCAAAAACAGAUGCUGAACAAGCUGAGUGCAAAACACAACAGAAAAGUGGCAA .....((((...((....)).......(((((..(((((....(.(((.....)))).(((((.......))))).))))).((.....))......))))).....))))... ( -23.00) >consensus ACUCUCCACAGAUGUAAACAGUAAAAAUGUUGAAUGUUCUUCCGCCGCUUUAUGUGCAGCAUCAAAAACAGAUGCUGAACAAGCCGAGUGCAAAGCAGAAAAGAAAAGCGGAUG ..........(((((.((((.......))))..)))))......(((((((.(((.(((((((.......))))))).))).((.....)).............)))))))... (-22.82 = -22.90 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:49 2006