
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
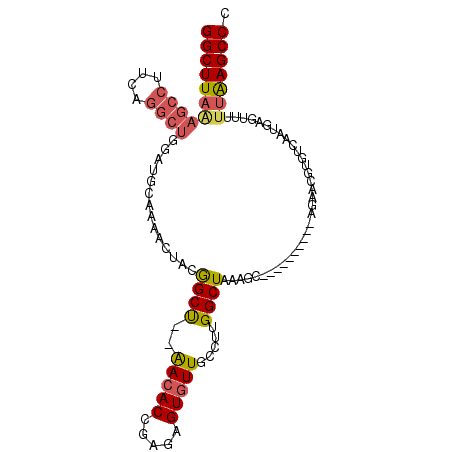
| Location | 3,894,192 – 3,894,285 |
| Length | 93 |
| Max. P | 0.796286 |

| Location | 3,894,192 – 3,894,285 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -18.05 |
| Energy contribution | -19.55 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3894192 93 + 27905053 GGCUUAAAGCCUUCAGGCUGGAUGCAAAACUACGGCU--AACACCGAAAGUGUUGCCUUGGCUAAAGC----------AGAACGUGUCAAUGAGUUUUUAAGCCC (((((((((((....)))).(((((........(((.--((((.(....))))))))...((....))----------.....))))).........))))))). ( -30.00) >DroSec_CAF1 17413 89 + 1 GGCUUAAAGCCUUCAGGCUGGAUGCAAAACUACGGCU--AACACCGAGAGUGUUGCCGUGGCUAAAGC----------AGAACGUGUCAAUGAGU----AAGCCC ((((((.((((....)))).(((((.....((((((.--(((((.....)))))))))))((....))----------.....)))))......)----))))). ( -33.20) >DroSim_CAF1 17597 93 + 1 GGCUUAAAGCCUUCAGGCUGGAUGCAAAACUACGGCU--AACACCGAGAGUGUUGCCUUGGCUAAAGC----------AGAACGUGUCAAUGAGUUUUUAAGCCC (((((((((((....)))).(((((........(((.--(((((.....))))))))...((....))----------.....))))).........))))))). ( -29.60) >DroEre_CAF1 16170 93 + 1 GGCUUAAAGCCUUCGGGCUGAAUGCAAAACUACGGCU--AACACCGAGAGUGUUGCCUCGGCUAAGGC----------AGAACGUGUCAAUGAGUUUUUGAGCCC (((((((((((((..((((((..((((.(((.(((..--....)))..))).)))).)))))))))))----------....(((....)))....)))))))). ( -34.00) >DroYak_CAF1 17514 92 + 1 GGCUUAAAGCCUUCGGGCUGGAUGCAAAACUACGGCU--AACACCAAGAGUGUUGC-UUGGCUAAAGC----------AGAACGUGUCAAUGAGUUUUUAAGCCC (((((((((((....)))).(((((........((..--....))....((.((((-((.....))))----------)).))))))).........))))))). ( -30.50) >DroPer_CAF1 30893 100 + 1 GGCUUAGGACAAACAGAGAGC-AGCAAA-GGGCAGCCGAGAAACUAAAAGUGUUGA---GGCUGAAGCUGAAGCUGGUGGAACGUGUCAAUGAGUUUUUAAGCCU ((((((((((...(((....(-(((...-...(((((..((.((.....)).))..---)))))..))))...)))......(((....))).).))))))))). ( -29.20) >consensus GGCUUAAAGCCUUCAGGCUGGAUGCAAAACUACGGCU__AACACCGAGAGUGUUGCCUUGGCUAAAGC__________AGAACGUGUCAAUGAGUUUUUAAGCCC (((((((((((....))))..............((((..(((((.....))))).....))))..................................))))))). (-18.05 = -19.55 + 1.50)


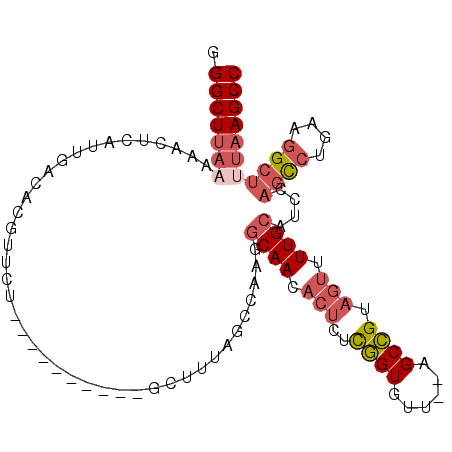
| Location | 3,894,192 – 3,894,285 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.62 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3894192 93 - 27905053 GGGCUUAAAAACUCAUUGACACGUUCU----------GCUUUAGCCAAGGCAACACUUUCGGUGUU--AGCCGUAGUUUUGCAUCCAGCCUGAAGGCUUUAAGCC .((((((((.................(----------((...(((...((((((((.....)))))--.)))...)))..)))....(((....))))))))))) ( -27.70) >DroSec_CAF1 17413 89 - 1 GGGCUU----ACUCAUUGACACGUUCU----------GCUUUAGCCACGGCAACACUCUCGGUGUU--AGCCGUAGUUUUGCAUCCAGCCUGAAGGCUUUAAGCC .(((((----(...............(----------((...(((.((((((((((.....)))))--.))))).)))..)))...((((....)))).)))))) ( -30.70) >DroSim_CAF1 17597 93 - 1 GGGCUUAAAAACUCAUUGACACGUUCU----------GCUUUAGCCAAGGCAACACUCUCGGUGUU--AGCCGUAGUUUUGCAUCCAGCCUGAAGGCUUUAAGCC .((((((((.................(----------((...(((...((((((((.....)))))--.)))...)))..)))....(((....))))))))))) ( -27.70) >DroEre_CAF1 16170 93 - 1 GGGCUCAAAAACUCAUUGACACGUUCU----------GCCUUAGCCGAGGCAACACUCUCGGUGUU--AGCCGUAGUUUUGCAUUCAGCCCGAAGGCUUUAAGCC (((((..(((((((.(((((((((..(----------(((((....))))))..)).....)))))--))..).))))))......)))))...(((.....))) ( -27.20) >DroYak_CAF1 17514 92 - 1 GGGCUUAAAAACUCAUUGACACGUUCU----------GCUUUAGCCAA-GCAACACUCUUGGUGUU--AGCCGUAGUUUUGCAUCCAGCCCGAAGGCUUUAAGCC .((((((((....((..((((((....----------(((...(((((-(.......))))))...--)))))).))).))......(((....))))))))))) ( -24.20) >DroPer_CAF1 30893 100 - 1 AGGCUUAAAAACUCAUUGACACGUUCCACCAGCUUCAGCUUCAGCC---UCAACACUUUUAGUUUCUCGGCUGCCC-UUUGCU-GCUCUCUGUUUGUCCUAAGCC .((((((...((.....((((.(......((((...((...(((((---.....((.....)).....)))))..)-)..)))-)....))))).))..)))))) ( -18.60) >consensus GGGCUUAAAAACUCAUUGACACGUUCU__________GCUUUAGCCAAGGCAACACUCUCGGUGUU__AGCCGUAGUUUUGCAUCCAGCCUGAAGGCUUUAAGCC .(((((((.........................................((((.(((..((((......)))).))).))))....((((....))))))))))) (-17.42 = -18.62 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:43 2006