
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,853,926 – 3,854,057 |
| Length | 131 |
| Max. P | 0.981557 |

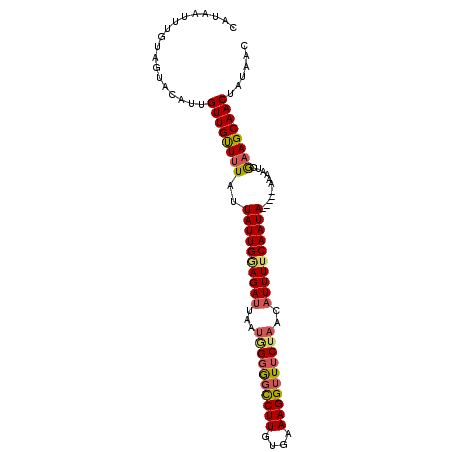
| Location | 3,853,926 – 3,854,028 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -18.27 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3853926 102 - 27905053 CAUAAUUUGUAGUGCAUUGUUGUUUUAUUAUUGCAGAUUAAUCGGGGCCUUGUGCAAGGUUUCUAACAUUUUCAAUAAAAUAAAAAUCGAAGCAACUAUAAC ......(((((((((.(((...(((((((((((.((((.....(..(((((....)))))..)....)))).)))))..))))))..))).)).))))))). ( -24.10) >DroSec_CAF1 11944 97 - 1 CAUAAGUUGUAGUACAUUGUUGCUUUAUUAUUGGAGAUUAAUGGGGGCCUUGUGAAAGGUUUCUAACAUUUUCAAUA-----AAAAUCGAAGCAACUAUAAC .....((((((((.......((((((.(((((((((((...(((..(((((....)))))..)))..))))))))))-----).....)))))))))))))) ( -29.71) >DroSim_CAF1 12423 97 - 1 CAUAAUUUGUAGUACAUUGUUGUUUUAUUAUUGGAGAUUAAUGGGGGCCUUGUGAAAGGUUUCUAACAUUUUCAAUA-----AAAAUCGAAGCAACUAUAAC ......(((((((....((((...((.(((((((((((...(((..(((((....)))))..)))..))))))))))-----).))....))))))))))). ( -26.20) >DroEre_CAF1 12290 97 - 1 CAUAAUUUGUAUCGUAUUGUUGUUCUAUUAUUGAAGAAUAGCAGAACUCUUGUGCAAGGUUCCUAACCUUUUCAAUA-----AAAAUCAAAGCAACUAUUAC .............(((..((((((...((((((((((......((((.(((....)))))))......)))))))))-----).......))))))...))) ( -16.30) >consensus CAUAAUUUGUAGUACAUUGUUGUUUUAUUAUUGGAGAUUAAUGGGGGCCUUGUGAAAGGUUUCUAACAUUUUCAAUA_____AAAAUCGAAGCAACUAUAAC ..................((((((((..((((((((((...((((((((((....))))))))))..))))))))))...........))))))))...... (-18.27 = -19.02 + 0.75)


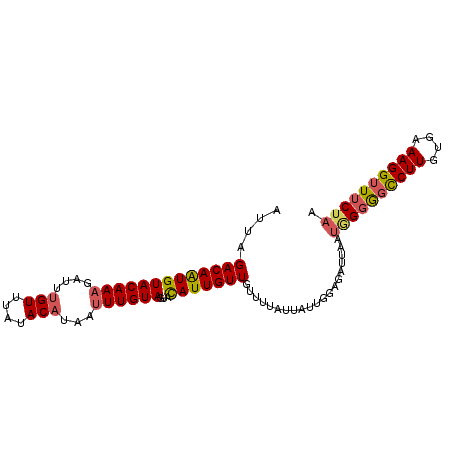
| Location | 3,853,962 – 3,854,057 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -16.64 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3853962 95 - 27905053 AUUAGACAGUGUACAAAGUUUUGUUUAUACAUAAUUUGUAGUGCAUUGUUGUUUUAUUAUUGCAGAUUAAUCGGGGCCUUGUGCAAGGUUUCUAA ....((((((((((.(((((.(((....))).)))))...))))))))))......................(..(((((....)))))..)... ( -24.80) >DroSec_CAF1 11975 95 - 1 AUUAGACAAUGUACAAAGAUUUGUUUAUACAUAAGUUGUAGUACAUUGUUGCUUUAUUAUUGGAGAUUAAUGGGGGCCUUGUGAAAGGUUUCUAA ....((((((((((...(((((((......)))))))...)))))))))).(((((....))))).....(((..(((((....)))))..))). ( -29.10) >DroSim_CAF1 12454 95 - 1 AUUAGACAAUGUACAAAGAUUUGUUUAUACAUAAUUUGUAGUACAUUGUUGUUUUAUUAUUGGAGAUUAAUGGGGGCCUUGUGAAAGGUUUCUAA ....((((((((((..((((((((....))).)))))...))))))))))....................(((..(((((....)))))..))). ( -26.30) >DroEre_CAF1 12321 94 - 1 CUUAGACAGUGUACAAAGAUACGUU-ACACAUAAUUUGUAUCGUAUUGUUGUUCUAUUAUUGAAGAAUAGCAGAACUCUUGUGCAAGGUUCCUAA ....(((..(((((((.((((((..-..........))))))((.((((((((((........)))))))))).))..)))))))..)))..... ( -25.40) >consensus AUUAGACAAUGUACAAAGAUUUGUUUAUACAUAAUUUGUAGUACAUUGUUGUUUUAUUAUUGGAGAUUAAUGGGGGCCUUGUGAAAGGUUUCUAA ....(((((((((((((....(((....)))...))))))...)))))))....................((((((((((....)))))))))). (-16.64 = -17.20 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:30 2006