
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,839,583 – 3,839,802 |
| Length | 219 |
| Max. P | 0.703648 |

| Location | 3,839,583 – 3,839,700 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -33.91 |
| Energy contribution | -32.45 |
| Covariance contribution | -1.46 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
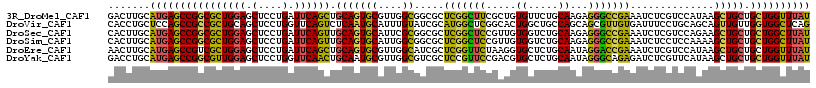
>3R_DroMel_CAF1 3839583 117 - 27905053 GACUUGCAUGAGCCGGCGCUGGAGCUCCUGAUUCAGCUGCAGUGCGUUGGCGGCGCUCGGCUUCGCUGUGUUCUGCAAGAGGGCCGAAAUCUCGUCCAUAAGCUGCUGCUGGUUUAU .......((((((((((((((((........)))))).(((((....((((((...((((((((..(((.....))).)).))))))....))).)))...))))).)))))))))) ( -45.90) >DroVir_CAF1 4075 117 - 1 CACCUGCUCCAGCCGCCGCUGCAGCUCCUGGUUCAGUCUCAAUGCAUUUGUAUCGCAUGGCUCGGCACUGGCUGCCAGCAGCGUUGUGAUUUCCUGCAGCAGUUGUUGGUGGCUCAG ....(((.(.(((((..((((.(((.....)))))))..).((((.........)))))))).))))(((((..(((((((((((((........))))).))))))))..)).))) ( -43.90) >DroSec_CAF1 4730 117 - 1 CACUUGCAUGAGCCGGCGCUGGAGCUCCUGAUUCAGUUGCAGUGCAUUCGCGGCGCUCGGCUCCGUUGUGGUCUGCAAGAGGGCCGAAAUCUCGUCCAGAAGCUGCUGCUGGCUUAU .......((((((((((((((.((((........)))).))))))....((((((((((((((..((((.....))))..)))))))..(((.....))).)).))))))))))))) ( -48.20) >DroSim_CAF1 4409 117 - 1 CACUUGCAUGAGCCGGCGCUGGAGCUCCUGAUUCAGUUGCAGUGCAUUGGCGGCGCUCGGCUCCGUUGUGGUCUGCAAGAGGGCCGAAAUCUCCUCCAAAAGCUGCUGCUGGCUUAU .......((((((((((((((((........)))))).(((((...((((.((.(.(((((((..((((.....))))..)))))))...).)).))))..))))).)))))))))) ( -50.30) >DroEre_CAF1 4193 117 - 1 AACUUGCAUGAGCCGUCGCUGGAGCUCCUGAUUCAGCUGCAGUGCGUUGGCAUCGCUCGGUUCUAAGGUGCUCUGCAAUAGGACCGAAAUCUCGUCCAUAAGCUGCUGCUGGUUUAU .......(((((((...((((((........)))))).(((((((..((((.....(((((((((...(((...))).)))))))))......).)))...)).))))).))))))) ( -39.70) >DroYak_CAF1 4093 117 - 1 GACCUGCAUGAGCCGGCGUUGGAGCUCCUGGUUCAACUGCAAUGCGUUGGCGUCGCUCCGUUCCGACGUGCUCUGCAAUAGGGCAGAGAUCUCGUUCAUAAGCUGCUGCUGGUUUAU .......((((((((((((((((.(....).)))))).(((..((..((((((((........)))))).((((((......))))))........))...))))).)))))))))) ( -42.00) >consensus CACUUGCAUGAGCCGGCGCUGGAGCUCCUGAUUCAGCUGCAGUGCAUUGGCGGCGCUCGGCUCCGAUGUGGUCUGCAAGAGGGCCGAAAUCUCGUCCAUAAGCUGCUGCUGGCUUAU .......((((((((((((((((.(....).)))))).(((((((....)).....(((((((.....((.....))...)))))))..............))))).)))))))))) (-33.91 = -32.45 + -1.46)
| Location | 3,839,700 – 3,839,802 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -41.21 |
| Consensus MFE | -28.30 |
| Energy contribution | -28.19 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
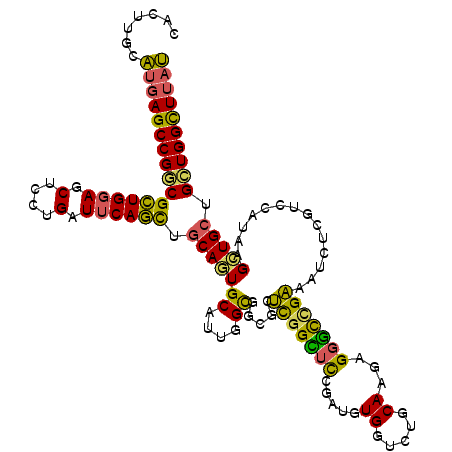
>3R_DroMel_CAF1 3839700 102 - 27905053 ---CGGGCCG---AAAUGGGAACGAGGUCGCAGCCGGCGCACCAGGAUGUCCUCGUUCCGAGAGAAGCGGGCCCUUACUACUUGUAACAUGGCCAGAAUUUUGGUCCG ---(((((((---((((.(((((((((..(((.((((....)).)).))))))))))))..........((((.((((.....))))...))))...))))))))))) ( -45.20) >DroSec_CAF1 4847 102 - 1 ---CGGGCCG---AAAUGGGAACGAGGUCGCAGCCGGCGCACCAGGAUGUCCUCGUUCCGGGAGAAGCGGGCCCUUACUACUUGUAACAUGGCCAGAAUUUUGGUCCG ---(((((((---((((.(((((((((..(((.((((....)).)).))))))))))))..........((((.((((.....))))...))))...))))))))))) ( -45.20) >DroSim_CAF1 4526 102 - 1 ---CGGGCCG---AAAUGGGAACGAGGUCGCAGCCGGCGCACCAGGAUGUCCUCGUUCCGGGAGAAGCGGGCCCUUACUACUUGUAACAUGGCCAGAAUUUUGGUCCG ---(((((((---((((.(((((((((..(((.((((....)).)).))))))))))))..........((((.((((.....))))...))))...))))))))))) ( -45.20) >DroEre_CAF1 4310 102 - 1 ---CGGGCCG---AAAUGGGAGCGAGGUCUCAGCCGGCGCACCAGGAUGUCCUCGUUUCGAGAGAAGCGGGCUCUAACUACUUGUAGCAUGGCCAGGAUUUUGGUCCG ---(((((((---((.(((..(((.(((....)))..))).)))....(((((((((((....))))))((((....(((....)))...)))))))))))))))))) ( -43.00) >DroYak_CAF1 4210 102 - 1 ---CGGGCCG---AAAUGGGAGCGAGGUCUAAGCCGGCGCACCAGGAUGUCCUCGUUCCGAGAGAAGCGGGUCCUAACUACUUGCAACAUGGCCAGGAUUUUAGUCCG ---..(((((---.....(((((((((.(....((((....)).))..).))))))))).......((((((.......))))))....))))).((((....)))). ( -39.60) >DroPer_CAF1 4463 108 - 1 AAGCCUGCUGUUACAUUGGGUGCGCGGCUUUAGGCGGCGGAUUAAAAUCUCCUCGUUCCGUGCAUAUCGAGCCCGGACAGAUUUCAGCAACGCUAGAACCUUUGCGCG ..((.(((((....(((..((.((.(((((.(.(((.((((...............)))))))...).))))))).)).)))..)))))..((..........)))). ( -29.06) >consensus ___CGGGCCG___AAAUGGGAACGAGGUCGCAGCCGGCGCACCAGGAUGUCCUCGUUCCGAGAGAAGCGGGCCCUUACUACUUGUAACAUGGCCAGAAUUUUGGUCCG ....(((((.......(.(((((((((.(....((((....)).))..).))))))))).)........)))))................((((((....)))))).. (-28.30 = -28.19 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:26 2006