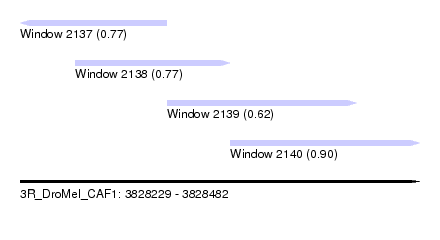
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,828,229 – 3,828,482 |
| Length | 253 |
| Max. P | 0.895231 |

| Location | 3,828,229 – 3,828,322 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3828229 93 - 27905053 CAAAAGAAGUGGACGCAGCUCAAGUAGACCACGAUCAAGGAAAUGAGCUGC------AAUCAAUUAAAAAUGUUCUGUUUUUUCGUUCUUUUCUAUAUC ....(((((.((((((((((((......((........))...))))))))------........((((((.....))))))..)))).)))))..... ( -21.90) >DroSec_CAF1 841 93 - 1 CAGAAGAAGUGGACGCAGCUCAAGUAGACCACGAUCAAGGAAAUGAGCUGC------AAUAAAUUUAAAAAGUUUUGUUAUUUCGUUCUUUUCUAUAUC ....(((((.((((((((((((......((........))...))))))))------((((((..........)))))).....)))).)))))..... ( -22.20) >DroSim_CAF1 589 93 - 1 CAGAAGAAGUGGACGCAGCUCAAGUAGACCACGAUCAAGGAAAUGAGCUGC------AAUCAAUUAAAGAAGUUUUGUUAUUUCGUUCUUUUCUAUAUC ....(((((.((((((((((((......((........))...))))))))------...........(((((......))))))))).)))))..... ( -24.60) >DroEre_CAF1 1701 99 - 1 CAGAAGAAGUGAACGCAGCUCAAAUAGACCACGAUCAAGGAAAUGAGCUGCGGAGCAAAUCAGUUAAAAAAUUGUUGUUAUAUCGUUCAUUUCUAUAUU ....((((((((((((((((((......((........))...))))))))..(((((..(((((....)))))))))).....))))))))))..... ( -30.40) >DroYak_CAF1 1668 99 - 1 CAGAAGAAGUGGACGCAGCUCAAAUAGACCACAAUCAAGGAAAUUAGCUAAGGAGCAAAUCAGUUUAAAAAGUUGUGUUAUUUCGUUCGCUUAUGUAUU ..(((((((((.((((((((.((((.((((........))......(((....)))...)).))))....)))))))))))))).)))........... ( -19.40) >consensus CAGAAGAAGUGGACGCAGCUCAAGUAGACCACGAUCAAGGAAAUGAGCUGC______AAUCAAUUAAAAAAGUUUUGUUAUUUCGUUCUUUUCUAUAUC ....(((((.((((((((((((......((........))...)))))))).........................(......))))).)))))..... (-15.44 = -15.92 + 0.48)


| Location | 3,828,264 – 3,828,362 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -27.79 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3828264 98 + 27905053 AUUGAUU------GCAGCUCAUUUCCUUGAUCGUGGUCUACUUGAGCUGCGUCCACUUCUUUUGGAGGCCAGAGAAUAGUGCCCACUUCGUGGUCGCCGGACAA .......------((((((((...((........))......))))))))((((..((((((.((...))))))))..(((.((((...)))).))).)))).. ( -32.90) >DroSec_CAF1 876 98 + 1 AUUUAUU------GCAGCUCAUUUCCUUGAUCGUGGUCUACUUGAGCUGCGUCCACUUCUUCUGGAAGCCAGAGAACAGUGCCCACUUCGUGGUCGCCGGACAG .......------((((((((...((........))......))))))))((((..((((.((((...))))))))..(((.((((...)))).))).)))).. ( -35.40) >DroSim_CAF1 624 98 + 1 AUUGAUU------GCAGCUCAUUUCCUUGAUCGUGGUCUACUUGAGCUGCGUCCACUUCUUCUGGAAGCCAGAGAACAGUGCCCACUUCGUGGUCGCCGGACAA .......------((((((((...((........))......))))))))((((..((((.((((...))))))))..(((.((((...)))).))).)))).. ( -35.40) >DroEre_CAF1 1736 104 + 1 ACUGAUUUGCUCCGCAGCUCAUUUCCUUGAUCGUGGUCUAUUUGAGCUGCGUUCACUUCUUCUGGAAGCCAGAGAACAGUGCACACUUCGUGAUCGCCGGACAG .......((.(((((((((((...((........))......))))))))((.(((((((.((((...)))))))..)))).))..............))))). ( -30.80) >DroYak_CAF1 1703 104 + 1 ACUGAUUUGCUCCUUAGCUAAUUUCCUUGAUUGUGGUCUAUUUGAGCUGCGUCCACUUCUUCUGGAAGCCAGAGAACAGUGCCCACUUUGUGGUCGCCGGACAG ........((((..((((((...((...))...))).)))...))))...((((..((((.((((...))))))))..(((.((((...)))).))).)))).. ( -27.60) >DroAna_CAF1 586 98 + 1 UUUAAUU------ACAGCUAAUUUCUCUGGUGAUUAUAUACUUGAGUUGUGUCCACUUUUUCUGGAAGCCGGAAAACAGCGCUCACUUUGUUGUUGCUGGCCAA .......------.((((......(((.((((......)))).)))..(((..(.((((((((((...)))))))).)).)..))).........))))..... ( -20.90) >consensus AUUGAUU______GCAGCUCAUUUCCUUGAUCGUGGUCUACUUGAGCUGCGUCCACUUCUUCUGGAAGCCAGAGAACAGUGCCCACUUCGUGGUCGCCGGACAA .............((((((((.......(((....)))....))))))))((((..((((.((((...))))))))..(((.((((...)))).))).)))).. (-27.79 = -27.10 + -0.69)


| Location | 3,828,322 – 3,828,442 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -43.35 |
| Consensus MFE | -28.74 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3828322 120 + 27905053 GAGGCCAGAGAAUAGUGCCCACUUCGUGGUCGCCGGACAAUCCAUGUGGAUAGCUGUAGCGGGCUAUCUGAGUAGCUUUCUUGGCGCCCUGCAGGUGCCGGUAUUUCUACUGGUUAUGAC ..((((((((((....(.((((...)))).)(((((((.((((....))))..((((((.((((((...(((.....))).)))).)))))))))).)))))..)))).))))))..... ( -43.90) >DroVir_CAF1 673 120 + 1 GAAGCCAGAGAAUAGCGCACGUUUCGUUGUCGCCGGCCAGUCCAUGUGGAUAGCCAUUGCCGGCUAUUUGUGCAGUUUUCUGGGCGCGUUUCAGGUGCCUGUUUUCCUGCUGGUCAUGGG ..(((..((((((((.((((.(..(((....(((((((.((((....)))).).....)))))).....((.(((....))).)))))....).))))))))))))..)))......... ( -40.70) >DroGri_CAF1 1457 120 + 1 GAAACCGGAGAAUAGCUCGCAUUUCGUUAUCGCUGGCCAAACUAUGUGGAUAGCCAUUGCUGGCUAUUUGUGCAGCUUUUUGGGCGCUCUUCAAGUGCCCGUCUUUCUGCUGGUGAUGGG ....((((.((.((((.........)))))).))))(((.((((.(..((((((((....))))))))..)((((.....((((((((.....)))))))).....))))))))..))). ( -42.70) >DroWil_CAF1 6786 120 + 1 GCGGCCAGAGAACCGAGCACAUUUUGUAGUUGCCGGUCAAUCCAUGUGGAUAGCCAUAGCCGGAUAUCUGUGCAGCUUUUUAGGAGCUCUUCAAGUACCAGUAUUCCUGCUGGUUAUGGC ...((((..(((..((((.(.......(((((((((...((((.(((((....)))))...))))..))).))))))......).)))))))....(((((((....)))))))..)))) ( -39.12) >DroMoj_CAF1 1530 120 + 1 GAAGCCGGAGAACAAAGCGCACUUCGUAGUUGCCGGGCAGUCUAUGUGGAUAGCCAUUGCCAGCUAUUUGUGCAGCUUUCUGGGCGCCCUUCAGGUGCCGGCUCUGCUGCUUGUCAUGAG .(((((((((...(((((((((...((((((((..(((.((((....)))).)))...).)))))))..)))).)))))...((((((.....))))))..)))))..))))........ ( -45.90) >DroAna_CAF1 644 120 + 1 GAAGCCGGAAAACAGCGCUCACUUUGUUGUUGCUGGCCAAACCAUGUGGGUAGCCAUCGCUGGCUACUUGAGCAGUUUUCUGGGUGCCCUUCAGGUGCCCGUUUUCCUGCUGGUUAUGGC ...(((((..(((((((.......))))))).)))))((((((....(((((((((....))))))))).(((((.....((((..((.....))..)))).....))))))))).)).. ( -47.80) >consensus GAAGCCAGAGAACAGCGCACACUUCGUUGUCGCCGGCCAAUCCAUGUGGAUAGCCAUUGCCGGCUAUUUGUGCAGCUUUCUGGGCGCCCUUCAGGUGCCCGUUUUCCUGCUGGUUAUGGC ...(((((........((((..............((.....))....((((((((......)))))))))))).......((((((((.....))))))))........)))))...... (-28.74 = -27.92 + -0.83)
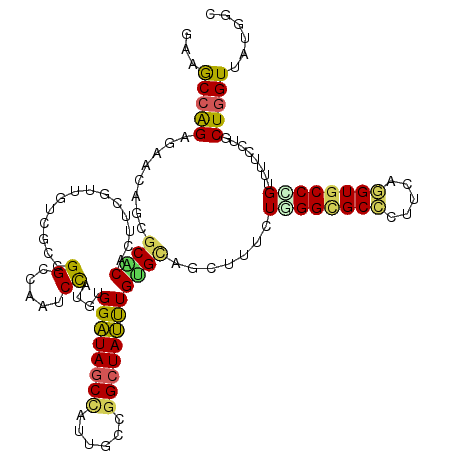

| Location | 3,828,362 – 3,828,482 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -40.25 |
| Consensus MFE | -27.95 |
| Energy contribution | -27.04 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3828362 120 + 27905053 UCCAUGUGGAUAGCUGUAGCGGGCUAUCUGAGUAGCUUUCUUGGCGCCCUGCAGGUGCCGGUAUUUCUACUGGUUAUGACCUAUGUAACAGCAUCUACGGCAUAUCAUCUGCAAACCCUA ....(((((((.(((((.((((((...(..((.......))..).)))).))(((((((((((....)))))))....)))).....))))))))))))(((.......)))........ ( -39.80) >DroVir_CAF1 713 120 + 1 UCCAUGUGGAUAGCCAUUGCCGGCUAUUUGUGCAGUUUUCUGGGCGCGUUUCAGGUGCCUGUUUUCCUGCUGGUCAUGGGCUACGUGCUUGUCUCCACGGCGUAUCAUCUGCAGACGAGU ..(((((((....((((.((((((........(((....)))(((((.......))))).........)))))).)))).)))))))(((((((.((.((.......)))).))))))). ( -39.20) >DroGri_CAF1 1497 120 + 1 ACUAUGUGGAUAGCCAUUGCUGGCUAUUUGUGCAGCUUUUUGGGCGCUCUUCAAGUGCCCGUCUUUCUGCUGGUGAUGGGCUACGUUUGCGUCUCAACGGCGUAUCAUUUACAGACGAGC .....(..((((((((....))))))))..)...((((...(((((((.....))))))).....((((..(((((((.((((((....)))......))).)))))))..)))).)))) ( -43.40) >DroWil_CAF1 6826 120 + 1 UCCAUGUGGAUAGCCAUAGCCGGAUAUCUGUGCAGCUUUUUAGGAGCUCUUCAAGUACCAGUAUUCCUGCUGGUUAUGGCCUAUGUAACGGCCUCGACGGCGUAUCAUCUACAGAUUAGC ....(((((((.(((.((((.(((...(((((((((((.....)))))......))).)))...))).))))(((..((((........))))..)))))).....)))))))....... ( -38.00) >DroMoj_CAF1 1570 120 + 1 UCUAUGUGGAUAGCCAUUGCCAGCUAUUUGUGCAGCUUUCUGGGCGCCCUUCAGGUGCCGGCUCUGCUGCUUGUCAUGAGCUACGUCAUUGCAUCCACAGCGUAUCAUCUACAGACGACU (((.(((((((.((.((.((.((((...((.(((((....(.((((((.....)))))).)....)))))....))..))))..)).)).)))))))))..(((.....))))))..... ( -38.80) >DroAna_CAF1 684 120 + 1 ACCAUGUGGGUAGCCAUCGCUGGCUACUUGAGCAGUUUUCUGGGUGCCCUUCAGGUGCCCGUUUUCCUGCUGGUUAUGGCAUAUGUUACGGCAUCGACGGCAUAUCACCUGCAGACAAGA ....((..(((.(((((.((..((.....((((((....))(((..((.....))..)))))))....))..)).)))))((((((..((.......)))))))).)))..))....... ( -42.30) >consensus UCCAUGUGGAUAGCCAUUGCCGGCUAUUUGUGCAGCUUUCUGGGCGCCCUUCAGGUGCCCGUUUUCCUGCUGGUUAUGGCCUACGUAACGGCAUCCACGGCGUAUCAUCUACAGACGAGA .(((((.((((((((......))))))))..((((.....((((((((.....)))))))).....))))....)))))..(((((.............)))))................ (-27.95 = -27.04 + -0.91)
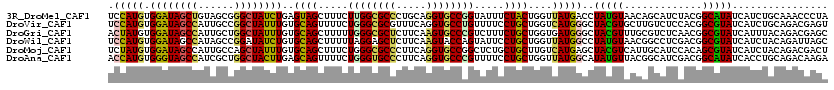

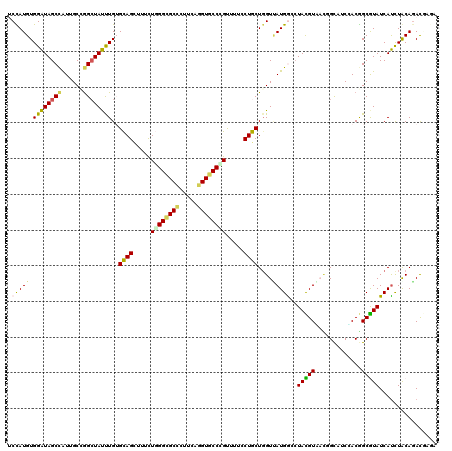
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:20 2006