
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,826,404 – 3,826,555 |
| Length | 151 |
| Max. P | 0.994922 |

| Location | 3,826,404 – 3,826,518 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -31.26 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3826404 114 - 27905053 UGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGUGAUAUGUGUGCUGCAGACACUCAAUUGAGGGGGAUUGACUCUGGAAAAAAGGAGCUUAAUCAUUAAAUACAU----ACGUUUACA ..(((((((...............)))))))((((.(((((((.........(((....)))...((((((((((........)))).))))))........)))----)))))))). ( -30.46) >DroSec_CAF1 54117 114 - 1 UGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGUGUGCUGCAGACACUCGAUUGAGGGGGAUUGACUCUGGAAAAAAGGAGCUUAAUCAUUAAAUACAU----ACGUUUACA ((((........)))).((((((((((((((((.....))))))))))....(((......))).((((((((((........)))).))))))..))))))...----......... ( -32.70) >DroSim_CAF1 58888 118 - 1 UGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGUGUGCUGCAGACACUCGAUUGAGGGGGAUUGACACUGGAAAAAAAGAGCUUAAUCAUUAAAUACAUAUGUACGUUUACA ..((((.......((........((((((((((.....))))))))))....(((....))))).(((((((.((........)).).)))))).............))))....... ( -29.40) >DroEre_CAF1 45508 113 - 1 UGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGCGUGCUGCAGGCACUCAAUUGAGGGGGAUUGACUCUGGAGAUAAGGAAUUUAAUCAUUAGAUA-----GUACUUAUAAA .(((((((((..(((........((((((((((.....))))))))))))).(((....)))...((((((.(((........)))..))))))....))))-----)))))...... ( -37.30) >DroYak_CAF1 60795 118 - 1 UGGUGCUGUUCAGCCAAUAUCUAUGCAGCACGCCACAUGCGUGCAGCAUGCACUCAAAUGGGGGAGAUUGACUCUGGAAAUAAAGAGUUUAAUCAUUGGAUAAAUAUGUACGUCUACA ..((((((((...(((((.(((((((.((((((.....)))))).))))...(((....))))))((((((((((........)))).)))))))))))...)))).))))....... ( -37.40) >consensus UGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGUGUGCUGCAGACACUCAAUUGAGGGGGAUUGACUCUGGAAAAAAGGAGCUUAAUCAUUAAAUACAU__GUACGUUUACA ((((........)))).((((((((((((((((.....))))))))))....(((....)))...((((((((((........)))).))))))..))))))................ (-31.26 = -30.50 + -0.76)



| Location | 3,826,440 – 3,826,555 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -19.04 |
| Energy contribution | -20.34 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3826440 115 + 27905053 UCCAGAGUCAAUCCCCCUCAAUUGAGUGUCUGCAGCACACAUAUCACGUGCUGCAUAAAUAUUGGCUGAACAGCACCAAAGACCGGUCAGAAGAGUCCUACAGCAAGGACAAGCA .....(((((((....(((....)))....((((((((.........)))))))).....))))))).....(((((.......))).......(((((......)))))..)). ( -30.90) >DroSec_CAF1 54153 115 + 1 UCCAGAGUCAAUCCCCCUCAAUCGAGUGUCUGCAGCACACAUAUCGCGUGCUGCAUAAAUAUUGGCUGAACAGCACCAAAGACCGAUCAGAAGAGUCCUACAGCAAGGACAAGCA .....(((((((....(((....)))....((((((((.(.....).)))))))).....))))))).....((............((....))(((((......)))))..)). ( -29.70) >DroSim_CAF1 58928 115 + 1 UCCAGUGUCAAUCCCCCUCAAUCGAGUGUCUGCAGCACACAUAUCGCGUGCUGCAUAAAUAUUGGCUGAACAGCACCAAAGACCGAUCAGAAGAGUCCUACAGCAAGGACAAACA .(((((((........(((....)))....((((((((.(.....).))))))))...)))))))((((.(.((......).).).))))....(((((......)))))..... ( -29.90) >DroEre_CAF1 45543 97 + 1 UCCAGAGUCAAUCCCCCUCAAUUGAGUGCCUGCAGCACGCAUAUCGCGUGCUGCAUAAAUAUUGGCUGAACAGCACCAAAGACCGAUCAGAACAGUC------------------ ....(((.........)))..(((.((((.((((((((((.....)))))))))).......((......)))))))))..................------------------ ( -27.10) >DroYak_CAF1 60835 95 + 1 UCCAGAGUCAAUCUCCCCCAUUUGAGUGCAUGCUGCACGCAUGUGGCGUGCUGCAUAGAUAUUGGCUGAACAGCACCAAAGACCGAUCAGAGCAA-------------------- ....(((.....))).....((((.((((((((.((((((.....)))))).))))......((......))))))))))...............-------------------- ( -26.00) >consensus UCCAGAGUCAAUCCCCCUCAAUUGAGUGUCUGCAGCACACAUAUCGCGUGCUGCAUAAAUAUUGGCUGAACAGCACCAAAGACCGAUCAGAAGAGUCCUACAGCAAGGACAA_CA ....(.(((.....................((((((((.........))))))))......((((.((.....)))))).))))..........(((((......)))))..... (-19.04 = -20.34 + 1.30)

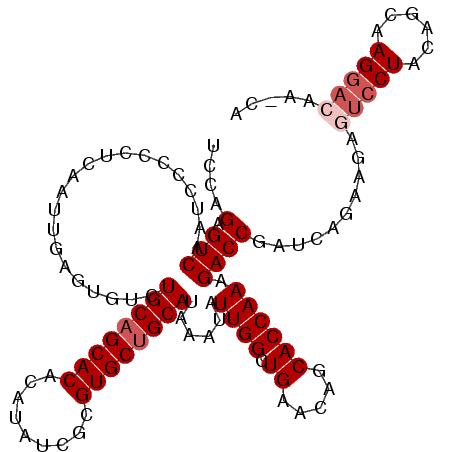
| Location | 3,826,440 – 3,826,555 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -29.22 |
| Energy contribution | -28.50 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3826440 115 - 27905053 UGCUUGUCCUUGCUGUAGGACUCUUCUGACCGGUCUUUGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGUGAUAUGUGUGCUGCAGACACUCAAUUGAGGGGGAUUGACUCUGGA .....(((((......)))))........((((...(..((.((.(((((..........(((((((((.......))))))))).........))))).)).))..)..)))). ( -35.51) >DroSec_CAF1 54153 115 - 1 UGCUUGUCCUUGCUGUAGGACUCUUCUGAUCGGUCUUUGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGUGUGCUGCAGACACUCGAUUGAGGGGGAUUGACUCUGGA ......(((......(((..((((((.((((((((.(((((........)))))......((((((((((.....)))))))))))))...)))))))))))..))).....))) ( -39.50) >DroSim_CAF1 58928 115 - 1 UGUUUGUCCUUGCUGUAGGACUCUUCUGAUCGGUCUUUGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGUGUGCUGCAGACACUCGAUUGAGGGGGAUUGACACUGGA ((((.(((((......)))))((((((((((((((.(((((........)))))......((((((((((.....)))))))))))))...))))).))))))...))))..... ( -39.80) >DroEre_CAF1 45543 97 - 1 ------------------GACUGUUCUGAUCGGUCUUUGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGCGUGCUGCAGGCACUCAAUUGAGGGGGAUUGACUCUGGA ------------------(((((.......)))))((..(.((.((((.(((........((((((((((.....))))))))))))).(((....)))..)))).).).)..)) ( -34.00) >DroYak_CAF1 60835 95 - 1 --------------------UUGCUCUGAUCGGUCUUUGGUGCUGUUCAGCCAAUAUCUAUGCAGCACGCCACAUGCGUGCAGCAUGCACUCAAAUGGGGGAGAUUGACUCUGGA --------------------....((((((((((((((.(((((((..((.......))..))))))).(((..((.((((.....)))).))..))).))))))))).)).))) ( -30.70) >consensus UG_UUGUCCUUGCUGUAGGACUCUUCUGAUCGGUCUUUGGUGCUGUUCAGCCAAUAUUUAUGCAGCACGCGAUAUGUGUGCUGCAGACACUCAAUUGAGGGGGAUUGACUCUGGA ....................(((((((((...(((.(((((........)))))......((((((((((.....)))))))))))))..)))...))))))............. (-29.22 = -28.50 + -0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:16 2006