
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,820,305 – 3,820,707 |
| Length | 402 |
| Max. P | 0.999509 |

| Location | 3,820,305 – 3,820,417 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -26.59 |
| Energy contribution | -26.02 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820305 112 + 27905053 CAGCGAUGCACUGCAUUUCCGUGUGAAAUGUUGUCAUGCCAGUGAGCUGGUGAGCUGGUGAGCUGGUGAGCUG--------GUGAGCUGGUGGGCUAGUGAGCUGGUCAAAGCCAAAAGC ((((.(((.((.(((((((.....))))))).)))))((((((..((..(....)..))..))))))..))))--------......((((.((((((....))))))...))))..... ( -43.20) >DroSec_CAF1 48075 120 + 1 CAGCGCUGCACUGCAUUUCCGUGUGAAAUGUUGUCGGGCCAGUGAGCUGGUGAGCUGGUGGGCUAGUGAGCUGGUGAGCUGGUGAGCUGGUGAGCUGGUGAGCUGGUCAAAGCCAAAAGC ((((.(((.((.(((((((.....))))))).)))))((((((..((..((..(((((....)))))..))..))..))))))..))))....(((..((.(((......)))))..))) ( -49.50) >DroSim_CAF1 52666 96 + 1 CAGCGCUGCACUGCAUUUCCGUGUGAAAUGUUGUCGGGCCAGUGAGCUGGUGAGCUGGUGCGCUAGUGAGCUG------------------------GUGAGCUGGUCAAAGCCAAAAGC ..(.((((.((.(((((((.....))))))).))).(((((((..((..((..(((((....)))))..))..------------------------))..)))))))..))))...... ( -37.10) >DroEre_CAF1 39476 96 + 1 CAGUGCUGCGCUGCAUUUCCGUGUGAAAUGUUGUCGGGCCAGUGAGCUGCUGAGCUGGUGAGCUGCUGAGCUG------------------------GAGAGCUGGUCAAAGCCAAAAUC ..(.(((.(((.(((((((.....))))))).).))(((((((...(((((.(((.((....))))).))).)------------------------)...)))))))..))))...... ( -32.90) >consensus CAGCGCUGCACUGCAUUUCCGUGUGAAAUGUUGUCGGGCCAGUGAGCUGGUGAGCUGGUGAGCUAGUGAGCUG________________________GUGAGCUGGUCAAAGCCAAAAGC ((((.(((.((.(((((((.....))))))).)))))(((((....)))))..))))...((((....))))...............................((((....))))..... (-26.59 = -26.02 + -0.56)

| Location | 3,820,345 – 3,820,457 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -29.94 |
| Energy contribution | -30.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820345 112 + 27905053 AGUGAGCUGGUGAGCUGGUGAGCUGGUGAGCUG--------GUGAGCUGGUGGGCUAGUGAGCUGGUCAAAGCCAAAAGCCAGCCUUCACUUCACUUGACCGCCAGCCAGUGAUGUUGCU ((..((((((((..(.(((((...(((((((((--------((....((((.((((((....))))))...))))...))))))..)))))))))).)..)))))))((....)))..)) ( -49.30) >DroSec_CAF1 48115 120 + 1 AGUGAGCUGGUGAGCUGGUGGGCUAGUGAGCUGGUGAGCUGGUGAGCUGGUGAGCUGGUGAGCUGGUCAAAGCCAAAAGCCAGCCUUCACUUCACUUGACCGCCAGCCAGCGAUGUUGCU .....((((....((((((((...(((((...(((((((((((....((((..((..(....)..))....))))...))))))..))))))))))...))))))))))))......... ( -51.50) >DroSim_CAF1 52706 96 + 1 AGUGAGCUGGUGAGCUGGUGCGCUAGUGAGCUG------------------------GUGAGCUGGUCAAAGCCAAAAGCCAGCCUUCACUUCACUUGACCGCCAGCCAGCGAUGUUGCU .((..(((((((..(.((((....(((((((((------------------------((....((((....))))...))))))..))))).)))).)..)))))))..))......... ( -38.90) >DroEre_CAF1 39516 94 + 1 AGUGAGCUGCUGAGCUGGUGAGCUGCUGAGCUG------------------------GAGAGCUGGUCAAAGCCAAAAUCCAGCCUUCACUUCACUUGACCGCCAGCCAGCGAUGUUG-- .......(((((.(((((((..(...((((.((------------------------(((.(((((.............)))))))))).))))...)..))))))))))))......-- ( -37.42) >consensus AGUGAGCUGGUGAGCUGGUGAGCUAGUGAGCUG________________________GUGAGCUGGUCAAAGCCAAAAGCCAGCCUUCACUUCACUUGACCGCCAGCCAGCGAUGUUGCU .....((((....(((((((..(.((((((..............((((....)))).(((((((((.............)))))..)))))))))).)..)))))))))))......... (-29.94 = -30.45 + 0.50)



| Location | 3,820,345 – 3,820,457 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820345 112 - 27905053 AGCAACAUCACUGGCUGGCGGUCAAGUGAAGUGAAGGCUGGCUUUUGGCUUUGACCAGCUCACUAGCCCACCAGCUCAC--------CAGCUCACCAGCUCACCAGCUCACCAGCUCACU ............((((((.(((..(((...((((..(((((.....((((.(((.....)))..))))..)))))))))--------..))).)))((((....))))..)))))).... ( -33.30) >DroSec_CAF1 48115 120 - 1 AGCAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUGGCUUUUGGCUUUGACCAGCUCACCAGCUCACCAGCUCACCAGCUCACCAGCUCACUAGCCCACCAGCUCACCAGCUCACU .........(((((((((.((.(.(((((.((((..(((((....(((......)))(((....)))...)))))))))..(((....)))))))).).)).)))))....))))..... ( -39.20) >DroSim_CAF1 52706 96 - 1 AGCAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUGGCUUUUGGCUUUGACCAGCUCAC------------------------CAGCUCACUAGCGCACCAGCUCACCAGCUCACU .........(((((..((((((...((..(((((..(((((...((((......))))....)------------------------))))))))).))..))).)))..)))))..... ( -33.20) >DroEre_CAF1 39516 94 - 1 --CAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUGGAUUUUGGCUUUGACCAGCUCUC------------------------CAGCUCAGCAGCUCACCAGCUCAGCAGCUCACU --.......(((((((((.(....(((...((...(((((((..((((......))))...))------------------------)))))..)).)))).))))).))))........ ( -31.90) >consensus AGCAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUGGCUUUUGGCUUUGACCAGCUCAC________________________CAGCUCACCAGCUCACCAGCUCACCAGCUCACU .......((((((((.....))).)))))(((((.((((((.((........)))))))).............................(((....(((......)))....)))))))) (-24.94 = -24.75 + -0.19)



| Location | 3,820,417 – 3,820,508 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 86.59 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -27.05 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820417 91 - 27905053 UCGAAGCUGUCGCUGUGUUAUGUCCGCAGUUGCAGCAGC--AGCACUUACACCAGCAACAUCACUGGCUGGCGGUCAAGUGAAGUGAAGGCUG ....((((.(((((......((.((((.(((((....))--))).....((((((........)))).)))))).)).....))))).)))). ( -32.10) >DroSec_CAF1 48195 91 - 1 UCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC--AGCACUUACAGCAGCAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUG .....(((((((((((...((.......)).))))))))--))).....((((....((.((((((((.....))).))))).))....)))) ( -35.20) >DroSim_CAF1 52762 91 - 1 UCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC--AGCACUUACAGCAGCAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUG .....(((((((((((...((.......)).))))))))--))).....((((....((.((((((((.....))).))))).))....)))) ( -35.20) >DroEre_CAF1 39572 79 - 1 GCGGAGCUGUUGCUGCAUUACGUACGCAGUUGCAGCAGCAG--------------CAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUG ((...((((((((((((..((.......)))))))))))))--------------).((.((((((((.....))).))))).))....)).. ( -32.70) >consensus UCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC__AGCACUUACAGCAGCAACAUCGCUGGCUGGCGGUCAAGUGAAGUGAAGGCUG .....(((((.((((((.......)))))).)))))................((((.((.((((((((.....))).))))).))....)))) (-27.05 = -27.68 + 0.63)



| Location | 3,820,457 – 3,820,548 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -18.40 |
| Energy contribution | -20.15 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820457 91 + 27905053 GGUGUAAGUGCU--GCUGCUGCAACUGCGGACAUAACACAGCGACAGCUUCGAAAUAAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUGGC ((((((((((((--((((((((....)))).))......(((....)))...........))))))))(((((...)))))))))))...... ( -28.70) >DroSec_CAF1 48235 91 + 1 GCUGUAAGUGCU--GCUGCUGCAACUGCGGACGUAACACAGCAACAGCUUCGAAAUAAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUCGC ((((((((((((--(((((((....(((....)))...)))))...((((......))))))))))))))))))................... ( -31.90) >DroSim_CAF1 52802 91 + 1 GCUGUAAGUGCU--GCUGCUGCAACUGCGGACGUAACACAGCAACAGCUUCGAAAUAAGUGCAACACUUGCAGUUUCUGUAUGCACUUUUCGC (((((..(((.(--((..((((....))))..))).)))....)))))..(((((..((((((.....(((((...)))))))))))))))). ( -28.80) >DroEre_CAF1 39610 80 + 1 ------------CUGCUGCUGCAACUGCGUACGUAAUGCAGCAACAGCUCCGCAA-AAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUGGC ------------(((.(((((((..(((....))).))))))).))).....(((-(((((((.....(((((...))))))))))))))).. ( -30.10) >consensus GCUGUAAGUGCU__GCUGCUGCAACUGCGGACGUAACACAGCAACAGCUUCGAAAUAAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUCGC .....((((((...((.((((((((((((..........(((....)))..........)))))...)))))))....))..))))))..... (-18.40 = -20.15 + 1.75)



| Location | 3,820,457 – 3,820,548 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -25.06 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820457 91 - 27905053 GCCAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUUAUUUCGAAGCUGUCGCUGUGUUAUGUCCGCAGUUGCAGCAGC--AGCACUUACACC .........(((.........)))(((((((((.(.......)..(((((.((((((.......)))))).))))).))--)))))))..... ( -30.30) >DroSec_CAF1 48235 91 - 1 GCGAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUUAUUUCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC--AGCACUUACAGC (((.....))).........(((.(((((((((.(.......)..(((((.((((((.......)))))).))))).))--))))))).))). ( -33.90) >DroSim_CAF1 52802 91 - 1 GCGAAAAGUGCAUACAGAAACUGCAAGUGUUGCACUUAUUUCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC--AGCACUUACAGC .((((((((((((((...........))).))))))..)))))..(((((((((((...((.......)).))))))))--)))......... ( -31.60) >DroEre_CAF1 39610 80 - 1 GCCAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUU-UUGCGGAGCUGUUGCUGCAUUACGUACGCAGUUGCAGCAGCAG------------ ((.((((((((((((...........))).)))))))-))))....(((((((((((..((.......)))))))))))))------------ ( -31.10) >consensus GCCAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUUAUUUCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC__AGCACUUACAGC ((...((((((((((...........))).)))))))........(((((.((((((.......)))))).))))).)).............. (-25.06 = -25.12 + 0.06)



| Location | 3,820,469 – 3,820,588 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -33.46 |
| Energy contribution | -33.65 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820469 119 - 27905053 CUCUGUUCUGUUUUGCUAGAGCCAUUCGCUUUCGAGCUCCGCCAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUUAUUUCGAAGCUGUCGCUGUGUUAUGUCCGCAGUUGCAGCAGC- ....((((((......)))))).....((((((((((...))...((((((((((...........))).)))))))...)))))(((((.((((((.......)))))).))))))))- ( -33.60) >DroSec_CAF1 48247 119 - 1 CUCGGUUCUGUUUUGCUAGAGCCAUUCGCUCUUGAGCUACGCGAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUUAUUUCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC- ...(((((((......)))))))....(((....)))....((((((((((((((...........))).))))))..)))))..(((((.((((((.......)))))).)))))...- ( -39.10) >DroSim_CAF1 52814 119 - 1 CUCGGUUCUGUUUUGCUAGAGCCAUUCGCUUUCGGGCUACGCGAAAAGUGCAUACAGAAACUGCAAGUGUUGCACUUAUUUCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC- ...(((((((......)))))))....(((....)))....((((((((((((((...........))).))))))..)))))..(((((.((((((.......)))))).)))))...- ( -38.50) >DroEre_CAF1 39611 119 - 1 CCCCGUUCUGUUUUGCUAGAGCCAUUCGCUAUGGAGCUCCGCCAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUU-UUGCGGAGCUGUUGCUGCAUUACGUACGCAGUUGCAGCAGCA ..((((..((..(.((....)).)..))..))))((((((((.((((((((((((...........))).)))))))-))))))))))(((((((((..((.......))))))))))). ( -48.70) >consensus CUCGGUUCUGUUUUGCUAGAGCCAUUCGCUUUCGAGCUACGCCAAAAGUGCAUACAGAAACUGCAAGUGCUGCACUUAUUUCGAAGCUGUUGCUGUGUUACGUCCGCAGUUGCAGCAGC_ ...(((((((......))))))).((((((....)))........((((((((((...........))).))))))).....)))(((((.((((((.......)))))).))))).... (-33.46 = -33.65 + 0.19)



| Location | 3,820,508 – 3,820,628 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820508 120 + 27905053 AAUAAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUGGCGGAGCUCGAAAGCGAAUGGCUCUAGCAAAACAGAACAGAGCAAAACGCUCAUGUUUUGUGGCAGAAAGCGGGGGAAAAGU ...(((((((.....(((((...))))))))))))((.(((((((((....).....)))))).((...((((((((((((.....)))).)))))))).)).....)).))........ ( -37.60) >DroSec_CAF1 48286 120 + 1 AAUAAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUCGCGUAGCUCAAGAGCGAAUGGCUCUAGCAAAACAGAACCGAGCAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGC ....((((((.....(((((...)))))))))))(..(((...((((.(((((.....)))))..(((((((.....((((.....)))).))))))))))).....)))..)....... ( -37.80) >DroSim_CAF1 52853 120 + 1 AAUAAGUGCAACACUUGCAGUUUCUGUAUGCACUUUUCGCGUAGCCCGAAAGCGAAUGGCUCUAGCAAAACAGAACCGAGCAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGC ....((((((.....(((((...)))))))))))(..(((...((((.((((((...((.(((........))).))((((.....)))).)))))).)))).....)))..)....... ( -38.60) >DroEre_CAF1 39651 119 + 1 AA-AAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUGGCGGAGCUCCAUAGCGAAUGGCUCUAGCAAAACAGAACGGGGCAAAACGCGCACGUUUUGGGGCAGAAAGCGGGGGAAAAGU ((-(((((((.....(((((...)))))))))))))).((....((((...((.....((((........((((((((.((.....)).).))))))))))).....)).))))....)) ( -35.90) >DroYak_CAF1 52088 120 + 1 AAUAAGUGCUGCACUUGCACUUUCUGCAUGCACUUUUGGCGGAGCUCAAAAGCGAAUGGCUCUAGCAAAACAGAACGGAGCAACACGCUCAUGUUUUGGGGCAUAAAACGGGGGAAAAGU .....((((.......))))(..(((.((((..(((((..(((((.((........)))))))..)))))(((((((((((.....)))).)))))))..))))....)))..)...... ( -34.10) >consensus AAUAAGUGCAGCACUUGCAGUUUCUGUAUGCACUUUUGGCGGAGCUCAAAAGCGAAUGGCUCUAGCAAAACAGAACCGAGCAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGU .............(((((..(((((((...........((((((((...........)))))).))....(((((((((((.....)))).)))))))..))))))))))))........ (-28.06 = -28.98 + 0.92)



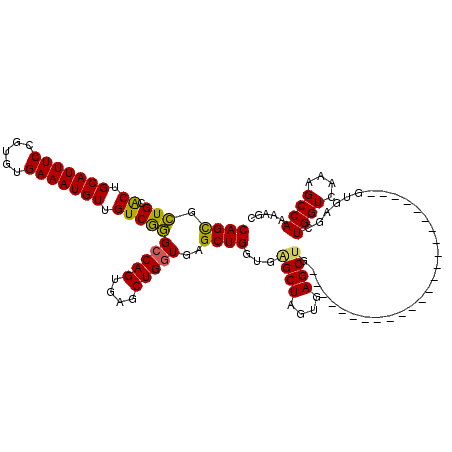
| Location | 3,820,548 – 3,820,668 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820548 120 + 27905053 GGAGCUCGAAAGCGAAUGGCUCUAGCAAAACAGAACAGAGCAAAACGCUCAUGUUUUGUGGCAGAAAGCGGGGGAAAAGUGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAA (((((((....).....)))))).((...((((((((((((.....)))).)))))))).))................((.(((.....(((..((....))..)))...))).)).... ( -40.60) >DroSec_CAF1 48326 120 + 1 GUAGCUCAAGAGCGAAUGGCUCUAGCAAAACAGAACCGAGCAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGCGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAA ...((((.(((((.....)))))..(((((((.....((((.....)))).)))))))))))................((.(((.....(((..((....))..)))...))).)).... ( -40.20) >DroSim_CAF1 52893 120 + 1 GUAGCCCGAAAGCGAAUGGCUCUAGCAAAACAGAACCGAGCAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGCGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAA ...((((.((((((...((.(((........))).))((((.....)))).)))))).))))................((.(((.....(((..((....))..)))...))).)).... ( -41.00) >DroEre_CAF1 39690 120 + 1 GGAGCUCCAUAGCGAAUGGCUCUAGCAAAACAGAACGGGGCAAAACGCGCACGUUUUGGGGCAGAAAGCGGGGGAAAAGUGGGCGCAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAA ....((((...((.....((((........((((((((.((.....)).).))))))))))).....)).))))....((.(((.(...(((..((....))..))).).))).)).... ( -39.40) >DroYak_CAF1 52128 120 + 1 GGAGCUCAAAAGCGAAUGGCUCUAGCAAAACAGAACGGAGCAACACGCUCAUGUUUUGGGGCAUAAAACGGGGGAAAAGUGGGCGAAAAGCGCGGAAAGGUCUGCGCGGAGUCGGCAAAA (((((.((........))))))).((....(((((((((((.....)))).)))))))..))................((.(((.....(((((((....)))))))...))).)).... ( -39.70) >consensus GGAGCUCAAAAGCGAAUGGCUCUAGCAAAACAGAACCGAGCAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGUGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAA ....(((....((.....((((...(((((((.....((((.....)))).))))))))))).....))..)))....((.(((.....(((((((....)))))))...))).)).... (-34.36 = -34.60 + 0.24)



| Location | 3,820,588 – 3,820,707 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -30.74 |
| Energy contribution | -31.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3820588 119 + 27905053 CAAAACGCUCAUGUUUUGUGGCAGAAAGCGGGGGAAAAGUGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAAAUGCAUUUGGGAAAAAACAGUGGAAAACCUUGCUA-GCGA .....((((..((((....))))...(((((((.....((.(((.....(((..((....))..)))...))).)).....(.((((.(........))))).)...))))))))-))). ( -32.50) >DroSec_CAF1 48366 119 + 1 CAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGCGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAAAUGCAUUGGGGAAAAGACAUUGGAAAACCUUGCUA-GCGA ......((((........)))).(..(((((((.....((.(((.....(((..((....))..)))...))).))...((((..((......))..))))......))))))).-.).. ( -33.80) >DroSim_CAF1 52933 119 + 1 CAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGCGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAAAUGCAUUGGGGAAAAGACAUUGGAAAACCUUGCUA-GCGA ......((((........)))).(..(((((((.....((.(((.....(((..((....))..)))...))).))...((((..((......))..))))......))))))).-.).. ( -33.80) >DroEre_CAF1 39730 119 + 1 CAAAACGCGCACGUUUUGGGGCAGAAAGCGGGGGAAAAGUGGGCGCAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAAAUGCAUUUGGGAAAAGACAUUGGAAAACCUUGCUG-GCGA (((((((....)))))))..((....(((((((.....((.(((.(...(((..((....))..))).).))).))...((((..(((....)))..))))......))))))).-)).. ( -37.40) >DroYak_CAF1 52168 120 + 1 CAACACGCUCAUGUUUUGGGGCAUAAAACGGGGGAAAAGUGGGCGAAAAGCGCGGAAAGGUCUGCGCGGAGUCGGCAAAAAUGCAUUUGGGAAAAGACAUUGGAAAACCUUGCUACGCGA ...(((.(((.(((((((.....))))))).)))....))).(((....(((((((....)))))))......(((((.((((..(((....)))..))))((....))))))).))).. ( -32.70) >consensus CAAAACGCUCAUGUUUUGGGGCAGAAAGCGGGGGAAAAGUGGGCGAAAAGCGUGGAAAGGUCUGCGCGGAGUCGGCAAAAAUGCAUUUGGGAAAAGACAUUGGAAAACCUUGCUA_GCGA (((((((....)))))))..((....(((((((.....((.(((.....(((((((....)))))))...))).))...((((..(((....)))..))))......)))))))..)).. (-30.74 = -31.18 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:08 2006