
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,819,782 – 3,819,916 |
| Length | 134 |
| Max. P | 0.961354 |

| Location | 3,819,782 – 3,819,879 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -20.40 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3819782 97 - 27905053 UUGCCUCUUUUGACACUGAAAAGUGCAAAGGCGCUGGCGGCGAAUUUUA-CUAGUUGCCAGUUGGCUAUGGGCGGCUAUG---------------------GGCUAUCGGGUCUGG-GUU ..((((..((((.((((....))))))))((((((((((((........-...)))))))))..)))..))))((((.((---------------------((((....)))))))-))) ( -34.00) >DroSec_CAF1 47576 84 - 1 UUGCCUCUUUUGACACUGAAAAGUGCACAGGCGCUGGCGGCGAAUUUUA-CUAG--GCCAGUUGGCUAU---CGGUUAUG-----------------------------GGUAUUG-GUU ..((((....((.((((....)))))).))))((..(((((........-....--))).))..)).((---((((....-----------------------------...))))-)). ( -23.20) >DroSim_CAF1 52164 84 - 1 UUGCCUCUUUUGACACUGAAAAGUGCAAAGGCGCUGGCGGCGAAUUUUA-CUAG--GCCAGUUGGCUAU---CGGGUAUG-----------------------------GGUCUUG-GUU .(((((((((((.((((....)))))))))).((..(((((........-....--))).))..))...---.)))))..-----------------------------.......-... ( -26.00) >DroEre_CAF1 38980 113 - 1 UUGCCCCUUUUGACACUGAAAAGUGCAAAGACGCUGGCGGCGAAUUUAA-CUGG--GCCAGUUGGCUAU---CGGCAAUGGGCUACGGGCUAUGGGCUAUGGGCUAUGG-GCUAUGGGGU ..((((((((((.((((....))))))))).(((.....))).......-...(--(((...(((((..---.(((.((((.(....).))))..)))...)))))..)-)))..))))) ( -43.90) >DroYak_CAF1 51401 79 - 1 UUGCCUCUUUUGACACUGAAAAGUGCAAAGACGCUGGCGGCGAAUUAAACCUGG--GCCAGUUGGCUAU---CGG-----------------------------------GCUGUU-GUU ..((((.(((((.((((....)))))))))..((..(((((.............--))).))..))...---.))-----------------------------------))....-... ( -24.52) >consensus UUGCCUCUUUUGACACUGAAAAGUGCAAAGGCGCUGGCGGCGAAUUUUA_CUAG__GCCAGUUGGCUAU___CGGCUAUG_____________________________GGUCUUG_GUU ..(((.((((((.((((....)))))))))).((..(((((...............))).))..)).......)))............................................ (-20.40 = -21.32 + 0.92)



| Location | 3,819,800 – 3,819,916 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -21.05 |
| Energy contribution | -20.85 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3819800 116 + 27905053 CAUAGCCGCCCAUAGCCAACUGGCAACUAG-UAAAAUUCGCCGCCAGCGCCUUUGCACUUUUCAGUGUCAAAAGAGGCAACACAAAAAUGCGGCAAAAGUCCCAGUCGAAAAUAAAA ....(((((..........(((((......-...........))))).(((((((((((....)))))....))))))...........)))))....................... ( -28.33) >DroSec_CAF1 47586 111 + 1 CAUAACCG---AUAGCCAACUGGC--CUAG-UAAAAUUCGCCGCCAGCGCCUGUGCACUUUUCAGUGUCAAAAGAGGCAACACAAAAAUGCGGCAAAAGUCCCAGUCAAAAAUAAAA ........---...(((..(((((--...(-.......)...))))).((((.((((((....)))))....).)))).............)))....................... ( -22.50) >DroSim_CAF1 52174 111 + 1 CAUACCCG---AUAGCCAACUGGC--CUAG-UAAAAUUCGCCGCCAGCGCCUUUGCACUUUUCAGUGUCAAAAGAGGCAACACAAAAAUGCGGCAAAAGUCCCAGUCGAAAAUAAAA ......((---((.(((....)))--....-........(((((....(((((((((((....)))))....))))))...........)))))..........))))......... ( -25.46) >DroEre_CAF1 39019 110 + 1 CAUUGCCG---AUAGCCAACUGGC--CCAG-UUAAAUUCGCCGCCAGCGUCUUUGCACUUUUCAGUGUCAAAAGGGGCAAC-CAAAAAUGCGGCAAAAGUCCCAGUCGAAAAUAAAA .(((..((---((.(((....)))--....-........(((((......(((((((((....)))))..))))((....)-)......)))))..........))))..))).... ( -27.60) >DroYak_CAF1 51410 107 + 1 -----CCG---AUAGCCAACUGGC--CCAGGUUUAAUUCGCCGCCAGCGUCUUUGCACUUUUCAGUGUCAAAAGAGGCAACACAAAAAUGCGGCAAAAGUCCCAGUCGAAAAUAAAU -----.((---((.(((..(((((--...(((.......))))))))..((((((((((....)))))..))))))))..........((.(((....))).))))))......... ( -26.50) >consensus CAUAGCCG___AUAGCCAACUGGC__CUAG_UAAAAUUCGCCGCCAGCGCCUUUGCACUUUUCAGUGUCAAAAGAGGCAACACAAAAAUGCGGCAAAAGUCCCAGUCGAAAAUAAAA ..............(((..(((((..................))))).(((((((((((....)))))....)))))).............)))....................... (-21.05 = -20.85 + -0.20)



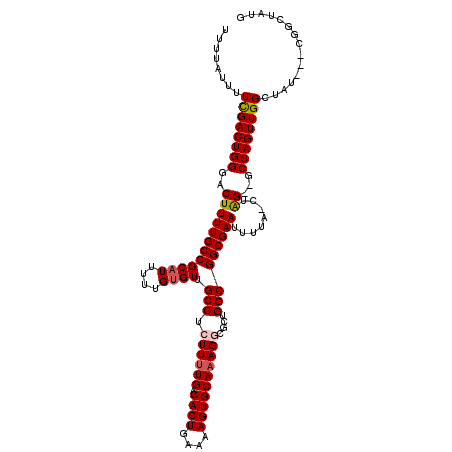
| Location | 3,819,800 – 3,819,916 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3819800 116 - 27905053 UUUUAUUUUCGACUGGGACUUUUGCCGCAUUUUUGUGUUGCCUCUUUUGACACUGAAAAGUGCAAAGGCGCUGGCGGCGAAUUUUA-CUAGUUGCCAGUUGGCUAUGGGCGGCUAUG .........(((((((....((((((((((....)))).(((.((((((.((((....))))))))))....))))))))).....-)))))))..(((((.(....).)))))... ( -38.60) >DroSec_CAF1 47586 111 - 1 UUUUAUUUUUGACUGGGACUUUUGCCGCAUUUUUGUGUUGCCUCUUUUGACACUGAAAAGUGCACAGGCGCUGGCGGCGAAUUUUA-CUAG--GCCAGUUGGCUAU---CGGUUAUG .........(((((((((..((((((((.((((.(((((.........))))).))))(((((....))))).))))))))..)).-...(--(((....)))).)---)))))).. ( -35.40) >DroSim_CAF1 52174 111 - 1 UUUUAUUUUCGACUGGGACUUUUGCCGCAUUUUUGUGUUGCCUCUUUUGACACUGAAAAGUGCAAAGGCGCUGGCGGCGAAUUUUA-CUAG--GCCAGUUGGCUAU---CGGGUAUG .......(((((((((....((((((((((....)))).(((.((((((.((((....))))))))))....))))))))).....-))))--(((....)))..)---)))).... ( -36.60) >DroEre_CAF1 39019 110 - 1 UUUUAUUUUCGACUGGGACUUUUGCCGCAUUUUUG-GUUGCCCCUUUUGACACUGAAAAGUGCAAAGACGCUGGCGGCGAAUUUAA-CUGG--GCCAGUUGGCUAU---CGGCAAUG .........(((((((..(.((((((((......(-(.....))(((((.((((....)))))))))......)))))))).....-..).--.)))))))((...---..)).... ( -32.70) >DroYak_CAF1 51410 107 - 1 AUUUAUUUUCGACUGGGACUUUUGCCGCAUUUUUGUGUUGCCUCUUUUGACACUGAAAAGUGCAAAGACGCUGGCGGCGAAUUAAACCUGG--GCCAGUUGGCUAU---CGG----- .........((((..((...((((((((......(((((......((((.((((....)))))))))))))..)))))))).....))..)--(((....)))..)---)).----- ( -34.80) >consensus UUUUAUUUUCGACUGGGACUUUUGCCGCAUUUUUGUGUUGCCUCUUUUGACACUGAAAAGUGCAAAGGCGCUGGCGGCGAAUUUUA_CUAG__GCCAGUUGGCUAU___CGGCUAUG ........((((((((..((((((((((((....)))).(((.((((((.((((....))))))))))....)))))))))........))...))))))))............... (-29.36 = -29.76 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:56 2006