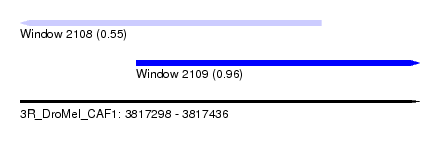
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,817,298 – 3,817,436 |
| Length | 138 |
| Max. P | 0.956670 |

| Location | 3,817,298 – 3,817,402 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.13 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.02 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
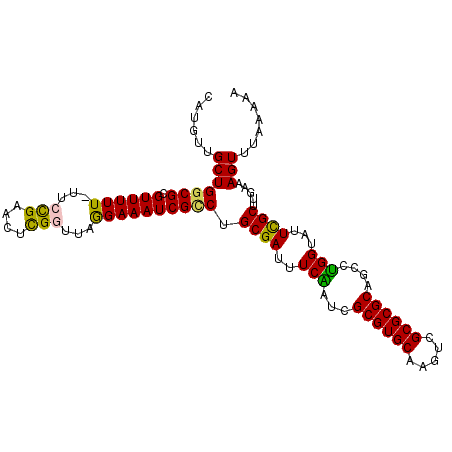
>3R_DroMel_CAF1 3817298 104 - 27905053 CAUGUUGCUGGCGCGUUUUU-UUCCGAACUCGGUUAGGAAAUCGCCUGCGAUUUCAAUCGCGUGCAAGUCGCGCGCAGUCUGGUAUUCGCUUGAAAGUUUAAAAA ...(((((.((((.((((((-(.(((....)))..))))))))))).))))).......((((((.....))))))..((.(((....))).))........... ( -31.60) >DroSec_CAF1 45271 104 - 1 CAUGUUGCUGGCGCGUUUUU-UUCCGAACUCGGUUAGGAAAUCGCCUGCGAUUUCAAUCGCGUGCAAGUCGCGCGCUUCCUGGUAUUCGCUUGAAAGUUUAAAAA ....((((.((((.((((((-(.(((....)))..))))))))))).))))(((((...((((((.....)))))).....(((....))))))))......... ( -31.00) >DroSim_CAF1 49788 104 - 1 CAUGUUGCUGGCGCGUUUUU-UUCCGAACUCGGUUAGGAAAUCGCCUGCGAUUUCAAUCGCGUGCAAGUCGCGCGCUACCUGGUAUUCGCUUGAAAGUUUAAAAA ....((((.((((.((((((-(.(((....)))..))))))))))).))))((((((.(((((((.....))))))(((...)))...).))))))......... ( -31.20) >DroEre_CAF1 36615 104 - 1 CAUGUUGCUGCCGCGUUUUU-UCCCGAGCUUGAUUGAGAAAUCGCCAGCGAUUUCGAUCGCGUGCAAGUCGCGCGCAGCUCGGUAUUCGCUGGAAAGUUUAAAAA ......(((.(((((..(..-..(((((((.((((((..(((((....)))))))))))((((((.....))))))))))))).)..))).))..)))....... ( -38.40) >DroYak_CAF1 49333 105 - 1 CAUGUUGCUGGCGCGUUUUUUUCGUGAGCUUGGUUAAGAAAUCGUCAGCGAUUUCGAUCGCGUGCAAGUCGCGCGCAGCUUGGUAUUUGCUUGAAAGUUGAAAAA ....(((((((((..((((..(((......)))..))))...)))))))))(((((((.((((((.....))))))(((.........))).....))))))).. ( -31.90) >consensus CAUGUUGCUGGCGCGUUUUU_UUCCGAACUCGGUUAGGAAAUCGCCUGCGAUUUCAAUCGCGUGCAAGUCGCGCGCAGCCUGGUAUUCGCUUGAAAGUUUAAAAA ......(((((((.((((((...(((....)))...)))))))))).((((..(((...((((((.....))))))....)))...)))).....)))....... (-25.74 = -25.02 + -0.72)


| Location | 3,817,338 – 3,817,436 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.36 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -13.39 |
| Energy contribution | -14.58 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3817338 98 + 27905053 GCACGCGAUUGAAAUCGCAGGCGAUUUCCUAACC-------GAGUUCGGAA-AAAAACGCGCCAGCAACAU--------GUGCACCAAAUGCACUUGCCACACCGCAACAAUAA--- ((.((((...(((((((....)))))))....((-------(....)))..-.....))))...((((...--------(((((.....)))))))))......))........--- ( -28.10) >DroSec_CAF1 45311 98 + 1 GCACGCGAUUGAAAUCGCAGGCGAUUUCCUAACC-------GAGUUCGGAA-AAAAACGCGCCAGCAACAU--------GUGCAGCAUAUGCACUUGCCACACAACAACAACAC--- ((..(((((....))))).((((.........((-------(....)))..-.......)))).))..((.--------(((((.....))))).)).................--- ( -26.77) >DroSim_CAF1 49828 98 + 1 GCACGCGAUUGAAAUCGCAGGCGAUUUCCUAACC-------GAGUUCGGAA-AAAAACGCGCCAGCAACAU--------GUGCAGCAUAUGCACUUGCCACACAACAACAACAC--- ((..(((((....))))).((((.........((-------(....)))..-.......)))).))..((.--------(((((.....))))).)).................--- ( -26.77) >DroEre_CAF1 36655 98 + 1 GCACGCGAUCGAAAUCGCUGGCGAUUUCUCAAUC-------AAGCUCGGGA-AAAAACGCGGCAGCAACAU--------GUGCAGCAAAUGCACUUGCCACACAGCAACAACGC--- ....(((...(....)(((((((.((((((....-------......))))-))...)))(((((......--------(((((.....))))))))))...)))).....)))--- ( -27.10) >DroYak_CAF1 49373 99 + 1 GCACGCGAUCGAAAUCGCUGACGAUUUCUUAACC-------AAGCUCACGAAAAAAACGCGCCAGCAACAU--------GUGCAGCAUAUGCACUUGCCACACAGCAACAACAC--- ((.((((...(((((((....)))))))......-------..(....)........))))...((((...--------(((((.....)))))))))......))........--- ( -22.00) >DroPer_CAF1 20381 106 + 1 GCACGCGAUUUAAAACGC--GCCAUUUCGUCAUACGCUCAUGUGUACGUAA-AGCGAAGCAACAACCACAUCGAAACACUUGUU------GCACUUGGUGCAC--CAACAACACCAA ................((--((((.(((((..((((..(....)..)))).-.)))))(((((((..............)))))------))...))))))..--............ ( -25.54) >consensus GCACGCGAUUGAAAUCGCAGGCGAUUUCCUAACC_______GAGUUCGGAA_AAAAACGCGCCAGCAACAU________GUGCAGCAUAUGCACUUGCCACACAGCAACAACAC___ ....(((((....))))).(((((..................................((....)).............(((((.....)))))))))).................. (-13.39 = -14.58 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:50 2006