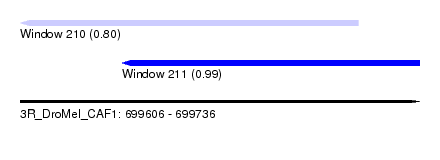
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 699,606 – 699,736 |
| Length | 130 |
| Max. P | 0.987013 |

| Location | 699,606 – 699,716 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.98 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -12.26 |
| Energy contribution | -16.07 |
| Covariance contribution | 3.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 699606 110 - 27905053 U-AUGGCUAGCAUACAAAUGGCCAUUGAUCAAUCUAUGGUACUUGUCCGUUCUAGAGCCCAACACUUUGUUUUUGAUCGUUAGGUCCAAAGCAAUUGA-------AAAUUGCCUACUU .-..(((((((..((((...(((((.((....)).)))))..))))..)))....))))...((..(((((((.((((....)))).))))))).)).-------............. ( -23.20) >DroSec_CAF1 118976 110 - 1 U-AUGGCUAGCAUAAAAAUGGCUUUUGAUCAAUCUUUGGUACUUGUCCGUUCUCGGACACAACACUUUGUGUUUGAUGGUUAAGUCCAAUGCAAUUGG-------CAUUUGGUUAUUU (-(((.....)))).((((((((..((.(((((..((((.(((((.(((...((((((((((....))))))))))))).)))))))))....)))))-------))...)))))))) ( -30.00) >DroSim_CAF1 121147 110 - 1 U-AUGGCUACCAUAUAAAUGGCCAUUGAUCAAUCUUUGGUACUUGUCCGUUCUCGGACACAACACUUUGUGUUUGAUGGUUAAGUCCAAUGAAAUUGG-------CAUUAGGUUAUUU (-((((...))))).((((((((..((.((((((.((((.(((((.(((...((((((((((....))))))))))))).))))))))).))..))))-------))...)))))))) ( -32.80) >DroYak_CAF1 129553 108 - 1 UCAUUGUCAGCAGAUUAAUGGCCAUUAAUCAAUUUUUGGU----------UCGCUGAACCAACACUCGGUAUUUGAGUGUAAUGCCCAAUGUAAUUGGGCAUUAGCAAUUGGCUAUGU .(((.((((((.(((((((....))))))).....(((((----------((...)))))))((((((.....))))))(((((((((((...)))))))))))))...)))).))). ( -36.10) >consensus U_AUGGCUAGCAUAUAAAUGGCCAUUGAUCAAUCUUUGGUACUUGUCCGUUCUCGGACACAACACUUUGUGUUUGAUGGUUAAGUCCAAUGCAAUUGG_______CAAUUGGCUAUUU ..(((((((.........)))))))...(((((..((((.(((((.(((((...((((((((....))))))))))))).)))))))))....))))).................... (-12.26 = -16.07 + 3.81)


| Location | 699,639 – 699,736 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.25 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -14.50 |
| Energy contribution | -16.00 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 699639 97 - 27905053 UGAAAUAC-------AUUGAUUGAUAUU-AUGGCUAGCAUACAAAUGGCCAUUGAUCAAUCUAUGGUACUUGUCCGUUCUAGAGCCCAACACUUUGUUUUUGAUC .(((((((-------((.(((((((...-(((((((.........)))))))..))))))).))).....(((..(((....)))...)))...))))))..... ( -20.80) >DroSec_CAF1 119009 97 - 1 CCCAAUAC-------AUUGAUUGAUAUU-AUGGCUAGCAUAAAAAUGGCUUUUGAUCAAUCUUUGGUACUUGUCCGUUCUCGGACACAACACUUUGUGUUUGAUG .......(-------(..(((((((...-..(((((.........)))))....)))))))..)).....((((((....)))))).(((((...)))))..... ( -24.70) >DroSim_CAF1 121180 97 - 1 CCCAAUAC-------AUUGAUUGAUAUU-AUGGCUACCAUAUAAAUGGCCAUUGAUCAAUCUUUGGUACUUGUCCGUUCUCGGACACAACACUUUGUGUUUGAUG .......(-------(..(((((((...-(((((((.........)))))))..)))))))..)).....((((((....)))))).(((((...)))))..... ( -28.70) >DroYak_CAF1 129593 95 - 1 CGAAAUACUAACCAUACUAAUUGAUAUUCAUUGUCAGCAGAUUAAUGGCCAUUAAUCAAUUUUUGGU----------UCGCUGAACCAACACUCGGUAUUUGAGU ..(((((((...........((((((.....))))))..(((((((....))))))).....(((((----------((...))))))).....))))))).... ( -19.40) >consensus CCAAAUAC_______AUUGAUUGAUAUU_AUGGCUAGCAUAUAAAUGGCCAUUGAUCAAUCUUUGGUACUUGUCCGUUCUCGGACACAACACUUUGUGUUUGAUG ..................(((((((....(((((((.........)))))))..)))))))..................((((((((((....)))))))))).. (-14.50 = -16.00 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:56 2006