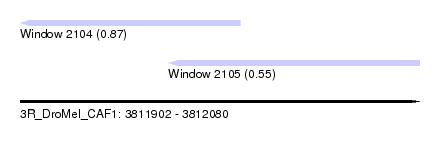
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,811,902 – 3,812,080 |
| Length | 178 |
| Max. P | 0.865543 |

| Location | 3,811,902 – 3,812,000 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -16.92 |
| Consensus MFE | -12.71 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3811902 98 - 27905053 UUUAAAGCCCCCUCAAUCGAAAA--CACAC------ACGAGGCAAUUUGUUAAUUUAUAUAAUUAUAAACUAAUAAGCCAAUGAACUUUAUUGGCUAUUGUUAUAU ..........((((.........--.....------..))))((((.(((((.((((((....)))))).)))))(((((((((...)))))))))))))...... ( -16.99) >DroSec_CAF1 40004 106 - 1 UUAAAAGCCCCCUCAAUCGAAAAUACACACACACACACGAGGCAAUUUGUUAAUUUAUAUAAUUAUGAAAUAAUAAGCCAAUGAACUUUAUUGGCUAUUGUUAUAU ........................................((((((.(((((.((((((....)))))).)))))(((((((((...))))))))))))))).... ( -16.80) >DroSim_CAF1 44511 100 - 1 UUAAAAGCCCCCUCAAUCGAAAAUACACACA------CAAGGCAAUUUGUUAAUUUAUAUAAUUAUGAAAUAAUAAGCCAAUGAACUUUAUUGGCUAUUGUUAUAU ...............................------...((((((.(((((.((((((....)))))).)))))(((((((((...))))))))))))))).... ( -16.80) >DroEre_CAF1 31563 96 - 1 UUAAAAUCCGCCUCAAUUGAAAA--C--AC------ACAAGGCAAUUUUUUAAUGUAUAUCAUUAGAAAAUAAUAAGUCAAUGAACUUUAUUGGAUAUUGUUAUAG .........((((....((....--)--).------...))))...(((((((((.....)))))))))(((((((.(((((((...)))))))...))))))).. ( -17.10) >consensus UUAAAAGCCCCCUCAAUCGAAAA__CACAC______ACAAGGCAAUUUGUUAAUUUAUAUAAUUAUAAAAUAAUAAGCCAAUGAACUUUAUUGGCUAUUGUUAUAU ........................................((((((.(((((.((((((....)))))).)))))(((((((((...))))))))))))))).... (-12.71 = -13.27 + 0.56)



| Location | 3,811,968 – 3,812,080 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -18.61 |
| Consensus MFE | -14.35 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3811968 112 - 27905053 UCCGACUAAGUCGAGAGGCAAGGCUUUUGGCCAAAACCAGAGACCAAAUUACAUACAAUUCAGUCCACAUAACUCGAACAUUUAAAGCCCCCUCAAUCGAAAA--CACAC------ACGA ..((((...)))).((((...(((((((((......)))(.(((..((((......))))..)))).................)))))).)))).........--.....------.... ( -19.40) >DroSec_CAF1 40070 119 - 1 UCCGACUAAGUCGAAAGGCAAGGCUUUUGGCCAAAACUAGAG-CCAAAUUACAUACAAUUCAGUCCACAUAACUCGAACAUUAAAAGCCCCCUCAAUCGAAAAUACACACACACACACGA ..(((....(((....)))..(((((((((((.......).)-)).............((((((.......))).)))....))))))).......)))..................... ( -19.70) >DroSim_CAF1 44577 114 - 1 UCCGACUAAGUCGAGAGGCAAGGCUUUUGGCCAAAACUAGAGACCAAAUUACAUACAAUUCAGUCCACAUAACUCGAACAUUAAAAGCCCCCUCAAUCGAAAAUACACACA------CAA ..((((...)))).((((...(((((((((.........(.(((..((((......))))..)))).(.......)....))))))))).)))).................------... ( -17.90) >DroEre_CAF1 31629 110 - 1 UCCGACUUGGUCGAGAGGCAAGGCUUUUGGACAAAAUUAGAGACCAAAUUACAUACAAUUCAGUCUACAUAACUCGAACAUUAAAAUCCGCCUCAAUUGAAAA--C--AC------ACAA ..((((...)))).(((((..((.((((((......(((.((((..((((......))))..))))...)))........)))))).))))))).........--.--..------.... ( -17.44) >consensus UCCGACUAAGUCGAGAGGCAAGGCUUUUGGCCAAAACUAGAGACCAAAUUACAUACAAUUCAGUCCACAUAACUCGAACAUUAAAAGCCCCCUCAAUCGAAAA__CACAC______ACAA ...((((..(((....)))..((.((((((......)))))).))................))))........((((...................)))).................... (-14.35 = -13.78 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:46 2006