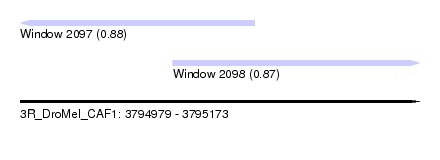
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,794,979 – 3,795,173 |
| Length | 194 |
| Max. P | 0.881594 |

| Location | 3,794,979 – 3,795,093 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -18.18 |
| Energy contribution | -19.70 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3794979 114 - 27905053 UGCGUCGGUUGUGGUGAGUGUGGAGUUGUUAAAAAUAAAACACACUGA----ACGCUGAUGAUCGAUGCGGGGGUCGCAAAGGUCCUACAAGG--AAAAAAGAGUAUCUGUCAGAUGCAG .((((((((..(((((((((((...((((.....))))..))))))..----.)))).)..)))))))).(((..(.....)..)))......--........(((((.....))))).. ( -37.20) >DroSec_CAF1 24323 115 - 1 UGCGUCGUUUGUGGUGAGUGUGGAGUUGUUAAAAAUAAAACACACUGA----UCGCUGGUGAUCGAUGCGGGGGUCGCAAAUGUCCAACGAGAA-AAAAAAGAGUAUCUGUCAGAUGCAG ((((((....(((((.((((((...((((.....))))..)))))).)----))))...((((.((((((((.((.....)).)))..(.....-......).))))).)))))))))). ( -34.60) >DroSim_CAF1 28802 116 - 1 UGCGUCGUUUGUGGUGAGUGUGGAGUUGUUAAAAAUAAAACACACUGA----ACGCUGGUGAUCGAUGCGGGGGUCGCCAAUGUCCUACGAGAAGAAAAAAGAGUAUCUGUCAGAUGCAG ((((((.((((((((.((((((...((((.....))))..)))))).)----(((.((((((((........)))))))).))).)))))))..((....((.....)).)).)))))). ( -35.30) >DroEre_CAF1 15485 97 - 1 UGCGUCGGUUGCGGUGGAGGUGUAGUUGUUAAAAAUAAAACACUCUGAAUAAAUGCUGAUGACCAAUGCGGG--------------------G---GAAAAGAGUAUCUGCCAUAUUGAG .((((.((((((((((...((.(((.((((........))))..))).))...))))).))))).))))((.--------------------(---((........))).))........ ( -22.80) >DroYak_CAF1 26584 117 - 1 UGCGUCGGUUGUGGUGGAUGUAGGGUUUUUAAAAAUAAAACACUCUGAAUAAACGCGGAUGACCAAUGCGGGGGUCGCAAAGGUCCUAUGAGG---GAAAAGAGUAUCGCCCAGAUGCAG (((((((((..(.(((.((.(((((((((((....))))).)))))).))...)))..)..))).....(((.(..((.....((((....))---)).....))..).))).)))))). ( -34.40) >consensus UGCGUCGGUUGUGGUGAGUGUGGAGUUGUUAAAAAUAAAACACACUGA____ACGCUGAUGAUCGAUGCGGGGGUCGCAAAGGUCCUACGAGG__AAAAAAGAGUAUCUGUCAGAUGCAG .((((((((((((.(.((((((...((((.....))))..)))))).).....)))....)))))))))..................................(((((.....))))).. (-18.18 = -19.70 + 1.52)

| Location | 3,795,053 – 3,795,173 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3795053 120 + 27905053 UUUUAUUUUUAACAACUCCACACUCACCACAACCGACGCAUCGGUGGCAAUUGCAAACAUUUAUACAAAUUGACGUUUAGCUGUCGCAGUUGUCACCUCGAAAGCCGCACCAAAGAUACA .....................................((...((((((((((((..(((.(((.((........)).))).))).)))))))))))).(....)..))............ ( -24.50) >DroSec_CAF1 24398 120 + 1 UUUUAUUUUUAACAACUCCACACUCACCACAAACGACGCAUCGGUGGCAAUUGCAAACAUUUAUAUAAAUUGACGUUUAGCUGUCGCAGUUGUCACCUCGAAAGCCGCACCAAAGAUACA .....................................((...((((((((((((....((((....)))).((((......)))))))))))))))).(....)..))............ ( -24.20) >DroSim_CAF1 28878 120 + 1 UUUUAUUUUUAACAACUCCACACUCACCACAAACGACGCAUCGGUGGCAAUUGCAAACAUUUAUACAAAUUGACGUUUAGCUGUCGCAGUUGUCACCUCGAAAGCCGCACCAAAGAUACA .....................................((...((((((((((((..(((.(((.((........)).))).))).)))))))))))).(....)..))............ ( -24.50) >DroEre_CAF1 15542 120 + 1 UUUUAUUUUUAACAACUACACCUCCACCGCAACCGACGCAUCAGUAGCAAUUGCAAACAUUUAUACAAAUUGACGUUUAGCUGUCGCAGUUGUCACCUCGAAAGCCGCACUAAAGAUUCA ....(((((((................((....))..((....((..(((((((..(((.(((.((........)).))).))).)))))))..))..(....)..))..)))))))... ( -16.20) >DroYak_CAF1 26661 120 + 1 UUUUAUUUUUAAAAACCCUACAUCCACCACAACCGACGCAUCAGUGGCAAUUGCAAACAUUUAUACAAAUUGACGUUUAGCUGUCGCAGUUGUCACCUCGAAAGCCGCACCAAAGAUACA .....................................((....(((((((((((..(((.(((.((........)).))).))).)))))))))))..(....)..))............ ( -20.70) >consensus UUUUAUUUUUAACAACUCCACACUCACCACAACCGACGCAUCGGUGGCAAUUGCAAACAUUUAUACAAAUUGACGUUUAGCUGUCGCAGUUGUCACCUCGAAAGCCGCACCAAAGAUACA .....................................((...((((((((((((..(((.(((.((........)).))).))).)))))))))))).(....)..))............ (-20.82 = -21.26 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:39 2006