
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,782,826 – 3,782,985 |
| Length | 159 |
| Max. P | 0.903385 |

| Location | 3,782,826 – 3,782,945 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -35.10 |
| Energy contribution | -36.54 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3782826 119 + 27905053 UAUGACUAAUUCCGGAGUCGGGCAGCGCCAGCGUGGGCAGCAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGCGACUAGGA-AAUUUGCAACAAAGUUGCCAUGGCCGGCUGGAAA .........((((((((((((((.(((((......))).)).(((((......)).))))))))))).(((...(((((((..(.-.........)..)))))))..)))...)))))). ( -42.80) >DroSec_CAF1 12263 119 + 1 UAUGACUAAUUCCGGAGUCGGGCAGCGCCAGUGUGGGCAGCAGGCCAAACUAAUGAGCCGUCCAACUUGUUUUGGGCGACUAGGA-AAUUUGCAACAAAGUUGCCUUGGCCGGCUGGAAA .........((((((.(((((((((((((......))).((((((((......)).)))((((((......))))))........-....)))......)))))).))))...)))))). ( -42.20) >DroSim_CAF1 15982 119 + 1 UAUGACUAAUUCCGGAGUCGGGCAGCGCCAGUGUGGGCAGCAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGCGACUAGGA-AAUUUGCAACAAAGUUGCCUUGGCCGGCUGGAAA .........((((((((((((((.(((((......))).)).(((((......)).))))))))))).(((..((((((((..(.-.........)..)))))))).)))...)))))). ( -43.40) >DroEre_CAF1 2614 120 + 1 CUUGACUAAAUCCGGAGUCGGGCAGCGCCAGUGUGGGCAGCAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGGGAUUAGGAAAACUUGCAACAAAGUUGCCAUGGCCAGCAAGAAA ((((.((...(((((((((((((.(((((......))).)).(((((......)).))))))))))))....))))((....((..(((((......))))).))....))))))))... ( -39.50) >DroYak_CAF1 15982 119 + 1 CAUGACUAAUUCCGGAGUCGGGCAGCGCCAGUGUGGGCAGCAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGG-GGACUGGAAAAAUGUGCAACAAAGUUGCCAUGGCCAGCAAGAAA .....((((..(..(((((((((.(((((......))).)).(((((......)).)))))))))))))..))))-.(.((((....(((.(((((...))))))))..)))))...... ( -42.20) >consensus UAUGACUAAUUCCGGAGUCGGGCAGCGCCAGUGUGGGCAGCAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGCGACUAGGA_AAUUUGCAACAAAGUUGCCAUGGCCGGCUGGAAA .........((((((.(((((((((((((......))).((((((((......)).)))((((((......)))))).............)))......)))))).))))...)))))). (-35.10 = -36.54 + 1.44)

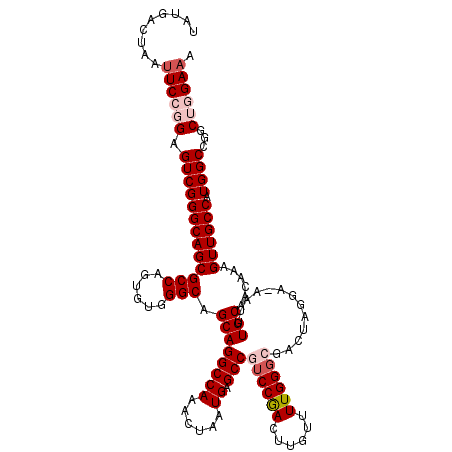

| Location | 3,782,866 – 3,782,985 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -35.84 |
| Energy contribution | -36.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3782866 119 + 27905053 CAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGCGACUAGGA-AAUUUGCAACAAAGUUGCCAUGGCCGGCUGGAAACCCGAUCUCGGUUCCUGUUUGAAGACGGGUUAGUUAGCUA ..(((((........((.(((((((......))))))).)).((.-(((((......))))).)).)))))((((((.((((((.((((((.......))).)))))))))..)))))). ( -40.60) >DroSec_CAF1 12303 119 + 1 CAGGCCAAACUAAUGAGCCGUCCAACUUGUUUUGGGCGACUAGGA-AAUUUGCAACAAAGUUGCCUUGGCCGGCUGGAAACCCGAUCUCGGUUCCUGUUUGAAGACGGGUUAGUUAGCUC ..((((((.......((.(((((((......))))))).))....-.....(((((...))))).))))))((((((.((((((.((((((.......))).)))))))))..)))))). ( -40.80) >DroSim_CAF1 16022 119 + 1 CAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGCGACUAGGA-AAUUUGCAACAAAGUUGCCUUGGCCGGCUGGAAACCCGAUCUCGGUUCCUGUUUGAAGACGGGUUAGUUAGCUC ..((((((.......((.(((((((......))))))).))....-.....(((((...))))).))))))((((((.((((((.((((((.......))).)))))))))..)))))). ( -40.50) >DroEre_CAF1 2654 120 + 1 CAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGGGAUUAGGAAAACUUGCAACAAAGUUGCCAUGGCCAGCAAGAAACCCGAUCUCGGUUCCUGUUUGAAGACGGGUUAGUUAGCUC ..(((..((((((((((((..((((......))))(((((..((....((((((((...)))((....))..)))))...))..))))))))))(((((....)))))))))))).))). ( -35.60) >DroYak_CAF1 16022 119 + 1 CAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGG-GGACUGGAAAAAUGUGCAACAAAGUUGCCAUGGCCAGCAAGAAACCCGAUCUCGGUUCCUGUUUGAAGACGGGUUAGUUAGCUC ..(((..(((((((((((((...(((..(((((..-.(.((((....(((.(((((...))))))))..)))))..)))))..).)).))))))(((((....)))))))))))).))). ( -36.00) >consensus CAGGCCAAACUAAUGAGCCGUCCGACUUGUUUUGGGCGACUAGGA_AAUUUGCAACAAAGUUGCCAUGGCCGGCUGGAAACCCGAUCUCGGUUCCUGUUUGAAGACGGGUUAGUUAGCUC ..(((((........((.(((((((......))))))).))..........(((((...)))))..)))))((((((.((((((.((((((.......))).)))))))))..)))))). (-35.84 = -36.04 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:16 2006