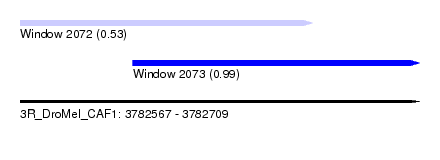
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,782,567 – 3,782,709 |
| Length | 142 |
| Max. P | 0.989182 |

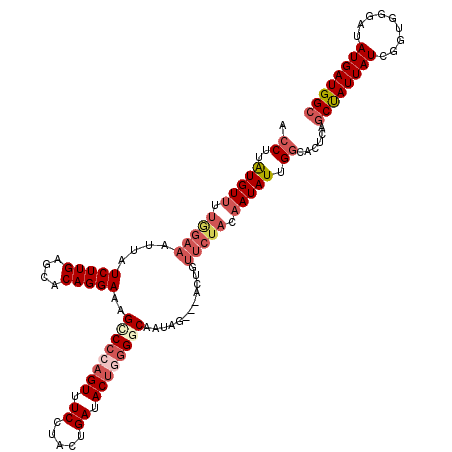
| Location | 3,782,567 – 3,782,671 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -15.92 |
| Energy contribution | -17.16 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3782567 104 + 27905053 CAGAUGUUUAAAAAUCACUAAACGCAUUACACUGACUAUAACCUUAUGUUUUGGAAAUUAUCUUGAGCACAGGAAAGUCGCAGUUUCCUACUGAUACUGGGGCA---------------- ..(.((((((........)))))))................(((..(((((.(((.....))).))))).)))...(((.((((.((.....)).)))).))).---------------- ( -19.30) >DroSec_CAF1 11994 115 + 1 CAGAUGUUUAAAAAUCACUAAAC-----ACACUGACCAUAACCUUGUGUUUUGGAAAUUAUCUUGAGCACAGGAAUGCCCCAGUUUCCUACUGAUACUGGGGCAAUUGACGACUGUUCUA (((.((((((........)))))-----)..)))........(((((((((.(((.....))).)))))))))..(((((((((.((.....)).)))))))))................ ( -35.60) >DroSim_CAF1 15708 117 + 1 CAGAUGUUUAAAAAUCACUAAACGCAUUACACUGACCAUAACCUUGUGUUUUGGAAAUUAUCUUGAGCACAGGAAAGACCCAGUUUCCUACUGAUACUGGGGCAAUUG---ACUGUUCUA (((..(((((........)))))((.................(((((((((.(((.....))).))))))))).....((((((.((.....)).)))))))).....---.)))..... ( -29.50) >DroEre_CAF1 2340 117 + 1 CAGAUGUUUAAAAAGCACUAAUCGCAUUACACUGGCCAUAACCUUAUGUUUUGGAAAUUAUCUUGAGUACAGGAAAGGCCCAGUAUCCAACGGAUACUAGGGCAAUAA---AUUGUUCUA .(((.(((((.......((..((.((..(((..((......))...)))..)))).....(((((....))))).))(((((((((((...))))))).))))..)))---))...))). ( -27.40) >DroYak_CAF1 15718 114 + 1 CAGAUGUUUAAAAAGCACUAAUCGCA---CACUGACCAUAGCCUUAUGUUUUAGAAAAUAUCUUGAGUACAGGAAAGCCCCAGUAUCCAACUGAUACCAGGGCAAUAU---AUUGUUCUA .((((((((.....((.......)).---..((((.((((....))))..)))).))))))))..((.((((....((((..(((((.....)))))..)))).....---.)))).)). ( -22.70) >consensus CAGAUGUUUAAAAAUCACUAAACGCAUUACACUGACCAUAACCUUAUGUUUUGGAAAUUAUCUUGAGCACAGGAAAGCCCCAGUUUCCUACUGAUACUGGGGCAAUAG___ACUGUUCUA (((.(((.....................)))))).(((.(((.....))).)))......(((((....)))))..((((((((.((.....)).))))))))................. (-15.92 = -17.16 + 1.24)

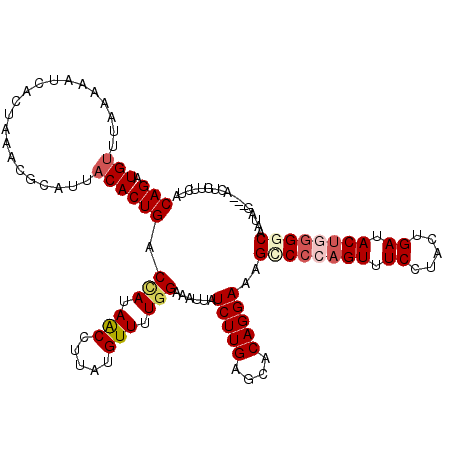
| Location | 3,782,607 – 3,782,709 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -21.88 |
| Energy contribution | -24.00 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3782607 102 + 27905053 ACCUUAUGUUUUGGAAAUUAUCUUGAGCACAGGAAAGUCGCAGUUUCCUACUGAUACUGGGGCA------------------AUAUUGGCACUCAGCUAUUAUCGGUGGGAUAUGAUGGC .(((..(((((.(((.....))).))))).)))...((((((...((((((((((((((((((.------------------......)).)))))....)))))))))))..)).)))) ( -32.90) >DroSec_CAF1 12029 117 + 1 ACCUUGUGUUUUGGAAAUUAUCUUGAGCACAGGAAUGCCCCAGUUUCCUACUGAUACUGGGGCAAUUGACGACUGUUCUACAAUAUUGGCAUUCAGCUAUUAUCG---GGAUAUGAUGGC ..(((((((((.(((.....))).)))))))))..(((((((((.((.....)).))))))))).....((.(((((((...(((.((((.....)))).))).)---))))).).)).. ( -38.10) >DroSim_CAF1 15748 114 + 1 ACCUUGUGUUUUGGAAAUUAUCUUGAGCACAGGAAAGACCCAGUUUCCUACUGAUACUGGGGCAAUUG---ACUGUUCUACAAUAUUGGCAUUCAGCUAUUAUCG---GGAUAUGAUGGC ..(((((((((.(((.....))).))))))))).....((((((.((.....)).)))))).....((---(.((((..........)))).)))((((((((..---....)))))))) ( -31.00) >DroEre_CAF1 2380 117 + 1 ACCUUAUGUUUUGGAAAUUAUCUUGAGUACAGGAAAGGCCCAGUAUCCAACGGAUACUAGGGCAAUAA---AUUGUUCUAGAAUAUUGGAACUCAGCCAUUAUCGGUGGAGUAUGAUGGC .((..(((((((((((....(((((....)))))...(((((((((((...))))))).)))).....---....))))))))))).))......(((((((((......).)))))))) ( -40.40) >DroYak_CAF1 15755 110 + 1 GCCUUAUGUUUUAGAAAAUAUCUUGAGUACAGGAAAGCCCCAGUAUCCAACUGAUACCAGGGCAAUAU---AUUGUUCUACAAUAUUG-------CCCAUUAUCGGUGGAGUAUGAUGGC ((((((((............(((((....)))))...........((((.((((((...(((((((((---..((.....))))))))-------)))..)))))))))).))))).))) ( -31.60) >consensus ACCUUAUGUUUUGGAAAUUAUCUUGAGCACAGGAAAGCCCCAGUUUCCUACUGAUACUGGGGCAAUAG___ACUGUUCUACAAUAUUGGCACUCAGCUAUUAUCGGUGGGAUAUGAUGGC .((..(((((.(((((....(((((....)))))..((((((((.((.....)).))))))))............))))).))))).))......((((((((.........)))))))) (-21.88 = -24.00 + 2.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:13 2006