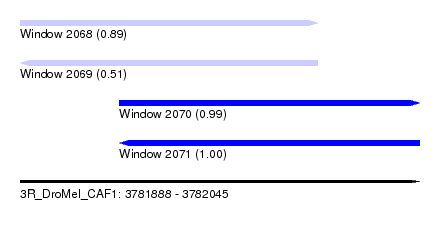
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,781,888 – 3,782,045 |
| Length | 157 |
| Max. P | 0.999950 |

| Location | 3,781,888 – 3,782,005 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -41.24 |
| Consensus MFE | -33.44 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3781888 117 + 27905053 AGCAAAGUUUUUCGUUGUCUGCCAGG-GGUUUUUCAGUCGAAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA--GAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAG ((((..((((..(((((((((.(.((-..(.(((..((((......))))...))).)..)).).)))))))).)..)--))).))))((((((((.((((.....)))))))))))).. ( -46.60) >DroSec_CAF1 11375 120 + 1 AGCAAAGUUUUUCGUUCUCUACCAGGGGGUUUUUCAGUCGAAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGAGAGAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAG .............(.(((((.((((...((((.((.((((......))))((.....))....)).))))..)))).))))).)...(((((((((.((((.....))))))))))))). ( -44.00) >DroSim_CAF1 15044 120 + 1 AGCAAAGUUUUUCGUUCUCUACCAGGGGGUUUUUCAGUCGAAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGAGAGAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAG .............(.(((((.((((...((((.((.((((......))))((.....))....)).))))..)))).))))).)...(((((((((.((((.....))))))))))))). ( -44.00) >DroEre_CAF1 1705 115 + 1 AGCAAAGUUUUUG-UUUC-UGCCAGG-GGUUUUUCAGUCGAAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA--GAUCUUUUGGUCUGGCACCAGGUCUUUUGGGCCUGGCCAG (((((.....)))-))((-(.((((.-.((((.((.((((......))))((.....))....)).))))..)))).)--)).....(((((.(((.((((.....))))))).))))). ( -36.20) >DroYak_CAF1 14984 116 + 1 AGCAAAGUUUUUG-UUUCCAGCCAGG-GGUUUUUCAGUCGAAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA--GAUCUUUUGGUCUUGCACCAGGUCUUUUGGGCCUGGCCAG .((..........-.......((((.-.((((.((.((((......))))((.....))....)).))))..))))((--((((....)))))))).(((((((.....))))))).... ( -35.40) >consensus AGCAAAGUUUUUCGUUCUCUGCCAGG_GGUUUUUCAGUCGAAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA__GAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAG .....................((((...((((.((.((((......))))((.....))....)).))))..))))...........(((((((((.((((.....))))))))))))). (-33.44 = -33.80 + 0.36)


| Location | 3,781,888 – 3,782,005 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -24.85 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3781888 117 - 27905053 CUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUC--UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUUCGACUGAAAAACC-CCUGGCAGACAACGAAAAACUUUGCU .((((((((((((.......))).))))))))).((((..--.....((((((((.((((....((((((..((......)))))))).....-.)))))))))))).......)))).. ( -41.64) >DroSec_CAF1 11375 120 - 1 CUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUCUCUCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUUCGACUGAAAAACCCCCUGGUAGAGAACGAAAAACUUUGCU .((((((((((((.......))).)))))))))...(.(((((.((((.(((((....)).)))((((((..((......)))))))).......)))).))))).)............. ( -37.60) >DroSim_CAF1 15044 120 - 1 CUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUCUCUCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUUCGACUGAAAAACCCCCUGGUAGAGAACGAAAAACUUUGCU .((((((((((((.......))).)))))))))...(.(((((.((((.(((((....)).)))((((((..((......)))))))).......)))).))))).)............. ( -37.60) >DroEre_CAF1 1705 115 - 1 CUGGCCAGGCCCAAAAGACCUGGUGCCAGACCAAAAGAUC--UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUUCGACUGAAAAACC-CCUGGCA-GAAA-CAAAAACUUUGCU ...(((((((......).))))))(((((.......(.((--(..(((....)))...))).).((((((..((......)))))))).....-.))))).-....-............. ( -23.20) >DroYak_CAF1 14984 116 - 1 CUGGCCAGGCCCAAAAGACCUGGUGCAAGACCAAAAGAUC--UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUUCGACUGAAAAACC-CCUGGCUGGAAA-CAAAAACUUUGCU (..((((((......((((((((.(..(((.(....).))--))))))..))))..........((((((..((......)))))))).....-))))))..)...-............. ( -25.30) >consensus CUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUC__UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUUCGACUGAAAAACC_CCUGGCAGAAAACGAAAAACUUUGCU .((((((((((((.......))).)))))))))............((((((..........((.....))..........))))))...........((((((...........)))))) (-24.85 = -25.77 + 0.92)

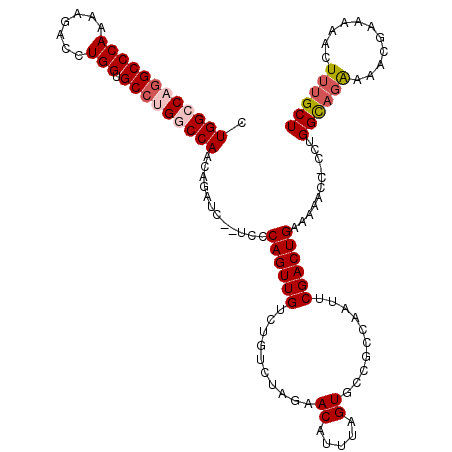
| Location | 3,781,927 – 3,782,045 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -39.48 |
| Energy contribution | -39.84 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3781927 118 + 27905053 AAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA--GAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAGGAGGCGCUGCCUGUCUUAAGAGCCCGAAAUUAGCACAACC ....((((((.....((.((((((........))))))--.))((.((((((((((.((((.....)))))))))))))).))..))))))(((.((((..........)))).)))... ( -44.90) >DroSec_CAF1 11415 120 + 1 AAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGAGAGAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAGGAGGCGCUGCCUGUCUUAAGAGCCCGAAAUUAGCACAACC ....((((((......(.((((((........)))))).)(.(((.((((((((((.((((.....)))))))))))))).))))))))))(((.((((..........)))).)))... ( -45.20) >DroSim_CAF1 15084 120 + 1 AAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGAGAGAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAGGAGGCGCUGCCUGUCUUAAGAGCCCGAAAUUAGCACAACC ....((((((......(.((((((........)))))).)(.(((.((((((((((.((((.....)))))))))))))).))))))))))(((.((((..........)))).)))... ( -45.20) >DroEre_CAF1 1742 118 + 1 AAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA--GAUCUUUUGGUCUGGCACCAGGUCUUUUGGGCCUGGCCAGGAGGCGCUGCCUGUCUUAAGGGCCCAAAAUUAGCACAACC ....((((((.....((.((((((........))))))--.))(((((((((.(((.((((.....))))))).)))))))))..)))))).(((.....)))................. ( -39.60) >DroYak_CAF1 15022 118 + 1 AAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA--GAUCUUUUGGUCUUGCACCAGGUCUUUUGGGCCUGGCCAGGAGGCGCUGCCUGUCUUAAGAGCCCGAAAUUAGCACAACC ..(((((..........(((((((((......((((((--((((....))))))...(((((((.....))))))))))).((((...))))))))..))))).........)).))).. ( -38.71) >consensus AAUUGGCGGCACUAAAUGUUCUAGACAGACAACUGGGA__GAUCUGUUGGCCAGGCACCAGGUCUUUUGGGCCUGGCCAGGAGGCGCUGCCUGUCUUAAGAGCCCGAAAUUAGCACAACC ....((((((........((((((........))))))..(.(((.((((((((((.((((.....)))))))))))))).))))))))))(((.((((..........)))).)))... (-39.48 = -39.84 + 0.36)


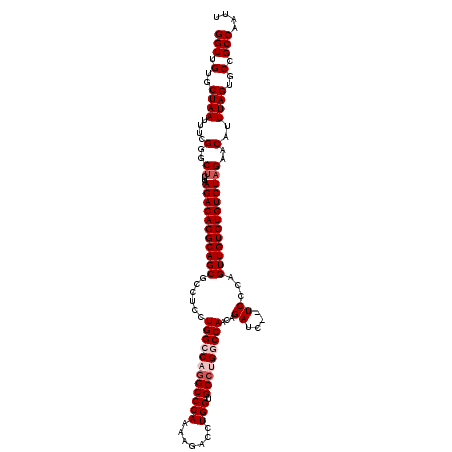
| Location | 3,781,927 – 3,782,045 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -46.64 |
| Consensus MFE | -43.58 |
| Energy contribution | -44.78 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.78 |
| SVM RNA-class probability | 0.999950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3781927 118 - 27905053 GGUUGUGCUAAUUUCGGGCUCUUAAGACAGGCAGCGCCUCCUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUC--UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUU (((.(..((((.....(..(((..(((((((((((......((((((((((((.......))).)))))))))...(...--...).)))))))))))))).)..))))..).))).... ( -50.60) >DroSec_CAF1 11415 120 - 1 GGUUGUGCUAAUUUCGGGCUCUUAAGACAGGCAGCGCCUCCUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUCUCUCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUU (((.(..((((.....(..(((..(((((((((((......((((((((((((.......))).)))))))))..((....))....)))))))))))))).)..))))..).))).... ( -51.10) >DroSim_CAF1 15084 120 - 1 GGUUGUGCUAAUUUCGGGCUCUUAAGACAGGCAGCGCCUCCUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUCUCUCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUU (((.(..((((.....(..(((..(((((((((((......((((((((((((.......))).)))))))))..((....))....)))))))))))))).)..))))..).))).... ( -51.10) >DroEre_CAF1 1742 118 - 1 GGUUGUGCUAAUUUUGGGCCCUUAAGACAGGCAGCGCCUCCUGGCCAGGCCCAAAAGACCUGGUGCCAGACCAAAAGAUC--UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUU (((.(..((((...((..(.....((((((((((((((....)))..((((((.......))).))).............--.....))))))))))).)..)).))))..).))).... ( -41.90) >DroYak_CAF1 15022 118 - 1 GGUUGUGCUAAUUUCGGGCUCUUAAGACAGGCAGCGCCUCCUGGCCAGGCCCAAAAGACCUGGUGCAAGACCAAAAGAUC--UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUU (((.(..((((.....(..(((..((((((((((((((....)))...(((((.......))).))..............--.....)))))))))))))).)..))))..).))).... ( -38.50) >consensus GGUUGUGCUAAUUUCGGGCUCUUAAGACAGGCAGCGCCUCCUGGCCAGGCCCAAAAGACCUGGUGCCUGGCCAACAGAUC__UCCCAGUUGUCUGUCUAGAACAUUUAGUGCCGCCAAUU (((.(..((((....(..((....(((((((((((......((((((((((((.......))).)))))))))...((....))...)))))))))))))..)..))))..).))).... (-43.58 = -44.78 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:11 2006