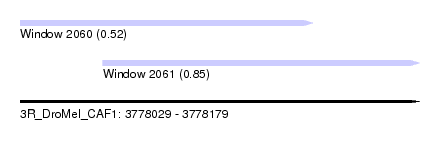
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,778,029 – 3,778,179 |
| Length | 150 |
| Max. P | 0.847971 |

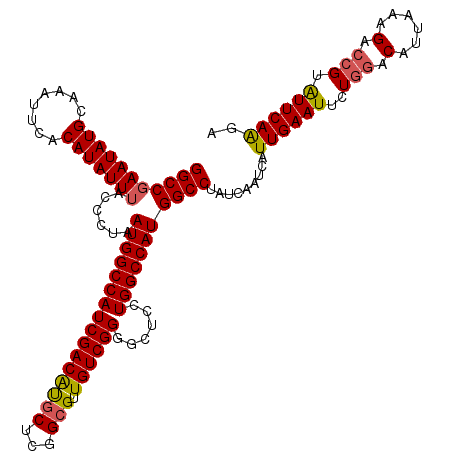
| Location | 3,778,029 – 3,778,139 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -36.39 |
| Energy contribution | -36.83 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3778029 110 + 27905053 GGUCGCCCGAGAGCCGUGGGUUGCU---------GGCUUUGGCCGAAUAUGCAAAUUCACAUAUUAA-CUUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUA (((.(((((.......))))).)))---------((((.((((((((((((........))))))..-......((((.(((((((((...))).))))))))))..)))))).)))).. ( -42.40) >DroSec_CAF1 7585 111 + 1 GGUCGCCCGAGAGCCGUGGGUUGCU---------GGCUUUGGCCGAAUAUGCAAAUUCACAUAUUUACCCUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUA (((.(((((.......))))).)))---------((((.((((((((((((........)))))))........((((.(((((((((...))).))))))))))...))))).)))).. ( -43.50) >DroSim_CAF1 11258 111 + 1 GGUCGCCCGAGAGCCGUGGGUUGCU---------GGCUUUGGCCGAAUAUGCAAAUUCACAUAUUUACCCUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUA (((.(((((.......))))).)))---------((((.((((((((((((........)))))))........((((.(((((((((...))).))))))))))...))))).)))).. ( -43.50) >DroYak_CAF1 9337 119 + 1 CGUCGC-CGAGAGACGUGGGUUGUUGGCAGUGGUGGCUGUGGCCGAAUAUGCAAAUUCACAUAUUUACCUUAAUGGCCAUCGACGCGCUCGGAGUUGUCGGUGCUCCUGGCCAUGGCCUA ((..((-(((..(((....))).)))))..))..(((((((((((((((((........)))))))........((.((((((((.((.....))))))))))..)).)))))))))).. ( -44.40) >consensus GGUCGCCCGAGAGCCGUGGGUUGCU_________GGCUUUGGCCGAAUAUGCAAAUUCACAUAUUUACCCUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUA .((.(((((.......))))).))..........(((..((((((((((((........)))))))........((((.(((((((((...))).))))))))))...)))))..))).. (-36.39 = -36.83 + 0.44)

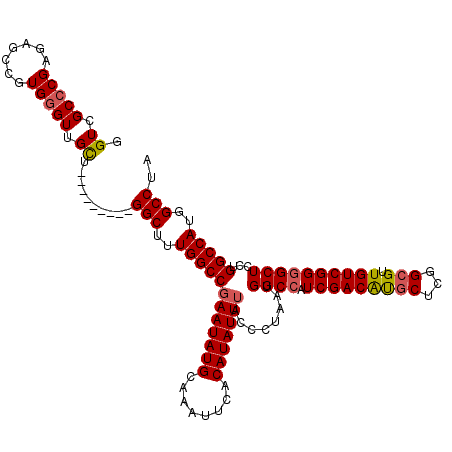
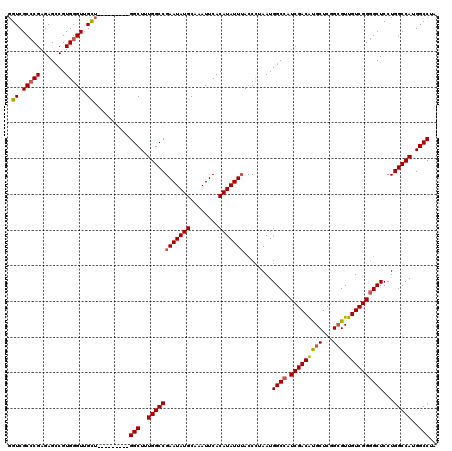
| Location | 3,778,060 – 3,778,179 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -31.42 |
| Energy contribution | -31.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3778060 119 + 27905053 GGCCGAAUAUGCAAAUUCACAUAUUAA-CUUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUAUCAAUCAUUGAAUUCUGGACAUUAAAGACCGUAUUCAAGA ((((.((((((........))))))..-....((((((..((((((((...))).)))))((....)))))))))))).........((((((..(((.(......).))).)))))).. ( -33.70) >DroSec_CAF1 7616 120 + 1 GGCCGAAUAUGCAAAUUCACAUAUUUACCCUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUAUCAAUCAUUGAAUUCUGGACAUUAAAGACCGUAUUCAAGA (((((((((((........)))))))......((((((..((((((((...))).)))))((....)))))))))))).........((((((..(((.(......).))).)))))).. ( -34.90) >DroSim_CAF1 11289 120 + 1 GGCCGAAUAUGCAAAUUCACAUAUUUACCCUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUAUCAAUCAUUGAAUUCUGGACAUUAAAGACCGUAUUCAAGA (((((((((((........)))))))......((((((..((((((((...))).)))))((....)))))))))))).........((((((..(((.(......).))).)))))).. ( -34.90) >DroYak_CAF1 9376 120 + 1 GGCCGAAUAUGCAAAUUCACAUAUUUACCUUAAUGGCCAUCGACGCGCUCGGAGUUGUCGGUGCUCCUGGCCAUGGCCUAUCAAUCAUUGAAUUCUGGACAUUAAAGAGAGUGUUCAGGA (((((((((((........)))))))......(((((((..((.((((.(((.....))))))))).)))))))))))..............(((((((((((......))))))))))) ( -42.40) >consensus GGCCGAAUAUGCAAAUUCACAUAUUUACCCUAAUGGCCAUCGACAUGCUCGGCGUUGUCGGGGCUCCUGGCCAUGGCCUAUCAAUCAUUGAAUUCUGGACAUUAAAGACCGUAUUCAAGA (((((((((((........)))))))......((((((((((((((((...))).))))))......))))))))))).........((((((..(((.(......).))).)))))).. (-31.42 = -31.67 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:01 2006