
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,777,489 – 3,777,641 |
| Length | 152 |
| Max. P | 0.863756 |

| Location | 3,777,489 – 3,777,601 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -22.89 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3777489 112 - 27905053 AGCACACUCGAGCUUUAAUUGUGUGUAUAAAUUACAUUUGCUGUAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAUAUCGCCAGAUGAUUUAGAUAGGACAC--------CAUGAU (((((((.(((.......))).)))).....(((((((..(((.(........).)))..))))))).)))(((((((..((((.....))))...)))))))...--------...... ( -26.90) >DroSec_CAF1 7072 97 - 1 ---------------UAAUUGUGUGUAUAAAUUACAUUUGCUGGAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAUAUCGCCAGAUGAUUUAGAUAGAACAC--------CAUGAU ---------------....((.((((.....(((((((..(((((........)))))..)))))))......(((((..((((.....))))...))))).))))--------)).... ( -25.20) >DroSim_CAF1 10708 112 - 1 AGCACACUCGAGCUUUAAUUGUGUGUAUAAAUUACACUUGCUGGAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAUAUCGCCAGAUGAUUUAGAUAGGACAC--------CAUGAU (((((((.(((.......))).)))).....(((((.(..(((((........)))))..).))))).)))(((((((..((((.....))))...)))))))...--------...... ( -29.00) >DroYak_CAF1 8819 114 - 1 AUCACAC------UUUAAUUAUGUGUAUAAAUUACAUUUGCUGGAUAAACAAUUUUAGUGGAUGUGGCGCUUCCUGUCAUAUCGCCAGAUGAUUUAGAUAGGAUACCAUGGUACCAUGAU .......------....((((((.((((.....(((((..(((((........)))))..)))))((....(((((((..((((.....))))...)))))))..))...)))))))))) ( -29.90) >consensus AGCACAC______UUUAAUUGUGUGUAUAAAUUACAUUUGCUGGAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAUAUCGCCAGAUGAUUUAGAUAGGACAC________CAUGAU .................((((((........(((((((..(((((........)))))..)))))))....(((((((..((((.....))))...)))))))...........)))))) (-22.89 = -23.32 + 0.44)

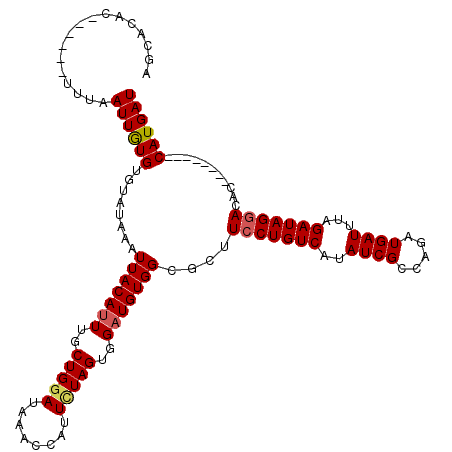

| Location | 3,777,521 – 3,777,641 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3777521 120 - 27905053 AAAUAGCAGCUAAAUUGCGGCGCGCAAACUCGAGCAUUUGAGCACACUCGAGCUUUAAUUGUGUGUAUAAAUUACAUUUGCUGUAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAU ..((((((((......)).((((((((....((((..(((((....)))))))))...))))))))............)))))).....((((....))))..((((((.....)))))) ( -30.90) >DroSec_CAF1 7104 103 - 1 AAAUAGCAGCUAAAUUGCGGCGCGCAAACUCGAGCAUU-----------------UAAUUGUGUGUAUAAAUUACAUUUGCUGGAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAU .....((.((......)).))((((........((((.-----------------.....))))........((((((..(((((........)))))..)))))))))).......... ( -29.80) >DroSim_CAF1 10740 120 - 1 AAAUAGCAGCUAAAUUGCGGCGCGCAAACUCGAGCAUUUGAGCACACUCGAGCUUUAAUUGUGUGUAUAAAUUACACUUGCUGGAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAU .....((((.......(((((((((((....((((..(((((....)))))))))...)))))))).....(((((.(..(((((........)))))..).))))))))...))))... ( -32.90) >DroYak_CAF1 8859 114 - 1 AAAUAGCAGCUGAAUUGCUGCGCGCAAACUCGAGCAUUUGAUCACAC------UUUAAUUAUGUGUAUAAAUUACAUUUGCUGGAUAAACAAUUUUAGUGGAUGUGGCGCUUCCUGUCAU .....(((((......)))))((((..................((((------.........))))......((((((..(((((........)))))..)))))))))).......... ( -32.40) >consensus AAAUAGCAGCUAAAUUGCGGCGCGCAAACUCGAGCAUUUGAGCACAC______UUUAAUUGUGUGUAUAAAUUACAUUUGCUGGAUAAACCAUUCUAGUGGAUGUGGCGCUUCCUGUCAU .....((.((......)).))((((........((((..((((........)))).....))))........((((((..(((((........)))))..)))))))))).......... (-25.94 = -26.88 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:59 2006