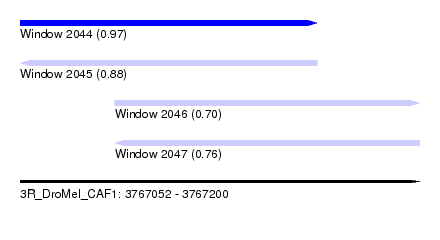
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,767,052 – 3,767,200 |
| Length | 148 |
| Max. P | 0.974988 |

| Location | 3,767,052 – 3,767,162 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.19 |
| Mean single sequence MFE | -36.74 |
| Consensus MFE | -10.93 |
| Energy contribution | -11.35 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
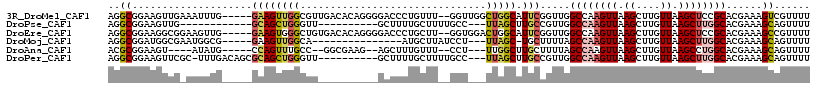
>3R_DroMel_CAF1 3767052 110 + 27905053 AGGCGGAAGUUGAAAUUUG-----GAAGUUGGCGUUGACACAGGGGACCCUGUUU--GGUUGGCUGGCAUUCGGUUGGCCAAGUUAAGCUUGUUAAGCUCCGCACGAAAGUCGUUUU ..(((((..((((......-----......(((.((((((((((....)))))((--(((..((((.....))))..)))))))))))))..))))..)))))((....))...... ( -37.94) >DroPse_CAF1 24381 92 + 1 AGGCGGAAGUUG------------GCAGCUGGGUU----------GCUUUUGCUUUUGCC---UUAGCUUGCCGUUGGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUU ((((((((((.(------------(((((...)))----------)))...)))))))))---)..((((..(((..((((((((.((....)).))))))))))).))))...... ( -37.30) >DroEre_CAF1 20479 110 + 1 AGGCGGAAGGCGGAAGUUG-----GAAGUGGGCUGUGACACAGGGGACCCUGCUU--GGUGGACUGGCAUUCGGUUGGCCAAGUUAAGCUUGUUAAGCUCCGCACGAAAGCCGUUUU .(((....((((((.(((.-----..((..((((.((((.((((....)))).((--(((.(((((.....))))).)))))))))))))..)).)))))))).)....)))..... ( -45.40) >DroMoj_CAF1 21945 93 + 1 AGGCGGAUGGCGAAUGGCG-----GAAGUUGGCA---------------AUGCUUAUCCU---UUAGC-UGCUUUUAGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUU ....((((((((..((((.-----...))))...---------------.))).))))).---..(((-(((((((.((((((((.((....)).))))))))..)))))))))).. ( -33.80) >DroAna_CAF1 18797 99 + 1 ACGCGGAAGU----AUAUG-----CCAGUUUGCC--GGCGAAG--AGCUUUGUUU--CCU---UUGGCUUGCUUUUAGCCAAGUUAAGCUUGUUAAGCCUGGCACGAAAGCAGUUUU ..((....))----...((-----(.....((((--(((....--(((...((((--..(---((((((.......)))))))..))))..)))..))).)))).....)))..... ( -29.40) >DroPer_CAF1 24860 103 + 1 AGGCGGAAGUUCGC-UUUGACAGCGCAGCUGGGUU----------GCUUUUGCUUUUGCC---UUAGCUUGCCGUUGGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUU ((((((((((..((-(.....)))(((((...)))----------))....)))))))))---)..((((..(((..((((((((.((....)).))))))))))).))))...... ( -36.60) >consensus AGGCGGAAGUUG_AAUUUG_____GAAGUUGGCUU__________GACUUUGCUU__GCU___UUAGCUUGCCGUUGGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUU ..((....................((((((((...............................))))).))).....((((((((.((....)).))))))))......))...... (-10.93 = -11.35 + 0.42)
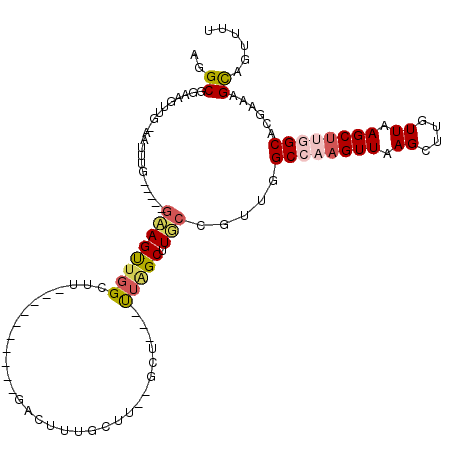

| Location | 3,767,052 – 3,767,162 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.19 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -8.77 |
| Energy contribution | -7.93 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3767052 110 - 27905053 AAAACGACUUUCGUGCGGAGCUUAACAAGCUUAACUUGGCCAACCGAAUGCCAGCCAACC--AAACAGGGUCCCCUGUGUCAACGCCAACUUC-----CAAAUUUCAACUUCCGCCU ...(((.....)))((((((((.....)))......((((.........))))((.....--..(((((....)))))......)).......-----............))))).. ( -23.22) >DroPse_CAF1 24381 92 - 1 AAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCCAACGGCAAGCUAA---GGCAAAAGCAAAAGC----------AACCCAGCUGC------------CAACUUCCGCCU ......((((((((((((((.((((.....))))))))))..)))).))))..(---(((..(((....(((----------......)))..------------...)))..)))) ( -24.60) >DroEre_CAF1 20479 110 - 1 AAAACGGCUUUCGUGCGGAGCUUAACAAGCUUAACUUGGCCAACCGAAUGCCAGUCCACC--AAGCAGGGUCCCCUGUGUCACAGCCCACUUC-----CAACUUCCGCCUUCCGCCU .....(((....(.((((((((.....))).....(((((.........)))))......--..(((((....)))))...............-----.....))))))....))). ( -25.80) >DroMoj_CAF1 21945 93 - 1 AAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCUAAAAGCA-GCUAA---AGGAUAAGCAU---------------UGCCAACUUC-----CGCCAUUCGCCAUCCGCCU ....(((((((...((((((.((((.....))))))))))..))))))-)....---.((((..((((---------------.((.......-----.)).))..)).)))).... ( -22.40) >DroAna_CAF1 18797 99 - 1 AAAACUGCUUUCGUGCCAGGCUUAACAAGCUUAACUUGGCUAAAAGCAAGCCAA---AGG--AAACAAAGCU--CUUCGCC--GGCAAACUGG-----CAUAU----ACUUCCGCGU .....((((....((((.(((......(((((...((((((.......))))))---.(.--...).)))))--....)))--))))....))-----))...----.......... ( -28.70) >DroPer_CAF1 24860 103 - 1 AAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCCAACGGCAAGCUAA---GGCAAAAGCAAAAGC----------AACCCAGCUGCGCUGUCAAA-GCGAACUUCCGCCU ......((((((((((((((.((((.....))))))))))..)))).))))..(---(((..(((....(((----------......))).((((.....)-)))..)))..)))) ( -28.90) >consensus AAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCCAACAGCAAGCCAA___AGC__AAGCAAAGGC__________AAGCCAACUGC_____CAAAUU_CAACUUCCGCCU ............(((..(((((.....)))))...(((((.........)))))..........................................................))).. ( -8.77 = -7.93 + -0.83)


| Location | 3,767,087 – 3,767,200 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
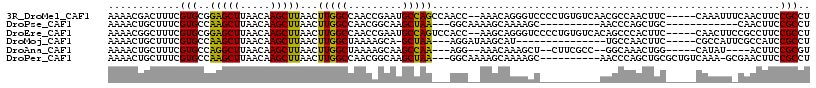
>3R_DroMel_CAF1 3767087 113 + 27905053 CAGGGGACCCUGUUU--GGUUGGCUGGCAUUCGGUUGGCCAAGUUAAGCUUGUUAAGCUCCGCACGAAAGUCGUUUUAAUCUCAAAAUUAAAACGUUUGCAAUUUUCGAGCGACU ((((....))))(((--(((..((((.....))))..))))))...((((.....)))).(((.((((((((((((((((......)))))))))......))))))).)))... ( -37.70) >DroPse_CAF1 24404 107 + 1 -----GCUUUUGCUUUUGCC---UUAGCUUGCCGUUGGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUUAAUCUCAAAAUUAAAACGUUUGCAAUUUUCAGGAGACU -----.((((((...((((.---..(((.(((..(((((((((((.((....)).)))))))).)))..)))((((((((......))))))))))).))))....))))))... ( -31.00) >DroGri_CAF1 26504 102 + 1 ---------AUGCUUGGCCU---UUAGC-UGCUUUUAGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUUAAUCUCAAAAUUAAAACGUUUGUAAUUUACCAGAAACU ---------....((((...---.....-(((((((.((((((((.((....)).))))))))..)))))))((((((((......))))))))............))))..... ( -25.40) >DroMoj_CAF1 21974 102 + 1 ---------AUGCUUAUCCU---UUAGC-UGCUUUUAGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUUAAUCUCAAAAUUAAAACGUUUGUAAUUUACCAGCAACU ---------.((((......---.....-(((((((.((((((((.((....)).))))))))..)))))))((((((((......))))))))..............))))... ( -26.20) >DroAna_CAF1 18826 108 + 1 AAG--AGCUUUGUUU--CCU---UUGGCUUGCUUUUAGCCAAGUUAAGCUUGUUAAGCCUGGCACGAAAGCAGUUUUAAUCUCAAAAUUAAAACGUUUGCAAUUUUCGAGAGACU ...--......((((--(..---((((((.......))))))((((.(((.....))).)))).((((((((((((((((......))))))))...)))...))))).))))). ( -25.20) >DroPer_CAF1 24894 107 + 1 -----GCUUUUGCUUUUGCC---UUAGCUUGCCGUUGGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUUAAUCUCAAAAUUAAAACGUUUGCAAUUUUCAGGAGACU -----.((((((...((((.---..(((.(((..(((((((((((.((....)).)))))))).)))..)))((((((((......))))))))))).))))....))))))... ( -31.00) >consensus _____G__UUUGCUU_UCCU___UUAGCUUGCUGUUAGCCAAGUUAAGCUUGUUAAGCUUGGCACGAAAGCAGUUUUAAUCUCAAAAUUAAAACGUUUGCAAUUUUCAAGAGACU ..........(((........................((((((((.((....)).))))))))....((((.((((((((......))))))))))))))).............. (-16.51 = -16.43 + -0.08)


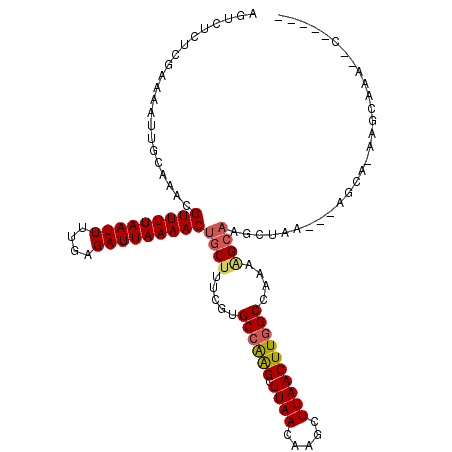
| Location | 3,767,087 – 3,767,200 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -15.91 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3767087 113 - 27905053 AGUCGCUCGAAAAUUGCAAACGUUUUAAUUUUGAGAUUAAAACGACUUUCGUGCGGAGCUUAACAAGCUUAACUUGGCCAACCGAAUGCCAGCCAACC--AAACAGGGUCCCCUG ...(((.(((((........((((((((((....))))))))))..))))).)))(((((.....)))))...(((((.............)))))..--...((((....)))) ( -28.32) >DroPse_CAF1 24404 107 - 1 AGUCUCCUGAAAAUUGCAAACGUUUUAAUUUUGAGAUUAAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCCAACGGCAAGCUAA---GGCAAAAGCAAAAGC----- ...............((....(((((((((....)))))))))((((((..((((.(((((..((((.....))))(((...))))))))..---))))))))))...))----- ( -27.80) >DroGri_CAF1 26504 102 - 1 AGUUUCUGGUAAAUUACAAACGUUUUAAUUUUGAGAUUAAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCUAAAAGCA-GCUAA---AGGCCAAGCAU--------- .(((..((((...........(((((((((....)))))))))((((((...((((((.((((.....))))))))))..))))))-.....---..)))))))..--------- ( -26.50) >DroMoj_CAF1 21974 102 - 1 AGUUGCUGGUAAAUUACAAACGUUUUAAUUUUGAGAUUAAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCUAAAAGCA-GCUAA---AGGAUAAGCAU--------- ...((((..............(((((((((....)))))))))((((((...((((((.((((.....))))))))))..))))))-.....---......)))).--------- ( -25.20) >DroAna_CAF1 18826 108 - 1 AGUCUCUCGAAAAUUGCAAACGUUUUAAUUUUGAGAUUAAAACUGCUUUCGUGCCAGGCUUAACAAGCUUAACUUGGCUAAAAGCAAGCCAA---AGG--AAACAAAGCU--CUU ((((((.(((((...(((...(((((((((....))))))))))))))))).)..))))).....(((((...((((((.......))))))---.(.--...).)))))--... ( -26.80) >DroPer_CAF1 24894 107 - 1 AGUCUCCUGAAAAUUGCAAACGUUUUAAUUUUGAGAUUAAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCCAACGGCAAGCUAA---GGCAAAAGCAAAAGC----- ...............((....(((((((((....)))))))))((((((..((((.(((((..((((.....))))(((...))))))))..---))))))))))...))----- ( -27.80) >consensus AGUCUCUCGAAAAUUGCAAACGUUUUAAUUUUGAGAUUAAAACUGCUUUCGUGCCAAGCUUAACAAGCUUAACUUGGCCAAAAGCAAGCUAA___AGCA_AAGCAAA__C_____ .....................(((((((((....)))))))))((((.....((((((.((((.....))))))))))....))))............................. (-15.91 = -16.13 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:47 2006