
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,764,615 – 3,764,716 |
| Length | 101 |
| Max. P | 0.999609 |

| Location | 3,764,615 – 3,764,716 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -17.93 |
| Energy contribution | -20.00 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3764615 101 + 27905053 AACCCAUGUAUAUGGUGCAGCUAUAUGUAUGUGCAGGUUGCACGUGUACUGGUACUGUGUACU-------------GGUAUUGGUACUGGGUACUGGUGUUGUUAUUACUUUUA .(((((.((((...((((((((...(((....)))))))))))..((((..((((...)))).-------------.))))..)))))))))...((((.......)))).... ( -31.80) >DroSec_CAF1 18131 110 + 1 AAUCCAUGUACAUGGUCCAGCUAUA----UGUGCAGGUUGCACGUGUACUAGUACUGUGUACUGUGCAGUGGUAUUGGUACUGGUACUGGGUACUGGUGUUGUUAUUACUUUUA ....(((((((((((((.((.((((----(((((.....))))))))))).).))))))))).))).(((((((.(..(((..((((...))))..)))..).))))))).... ( -38.70) >DroEre_CAF1 17829 83 + 1 ACCCCAUGUACAUGGUGCAGCUAUU----CGUGCAGGCUGCACUUGUACUGGUAC--------------------------UGCUGCUG-GUACUGGUGUUGUUAUUACUUUUA (((....(((((.(((((((((...----......)))))))))))))).(((((--------------------------(......)-))))))))................ ( -26.90) >DroYak_CAF1 20544 83 + 1 ACCCCAUGUACAUGAUGGAGCUCUU----UGUGCAGGCUGCACUGGUACUGGUGC--------------------------UAGUACUG-GUACUGGUGUUGUUAUUACUUUUA ......((((((.((......))..----))))))(((.((((..((((..(((.--------------------------...)))..-))))..)))).))).......... ( -28.20) >consensus AACCCAUGUACAUGGUGCAGCUAUA____UGUGCAGGCUGCACGUGUACUGGUAC__________________________UGGUACUG_GUACUGGUGUUGUUAUUACUUUUA ...(((.((((...((((((((.............))))))))..(((((((((((....................)))))))))))...)))))))................. (-17.93 = -20.00 + 2.06)

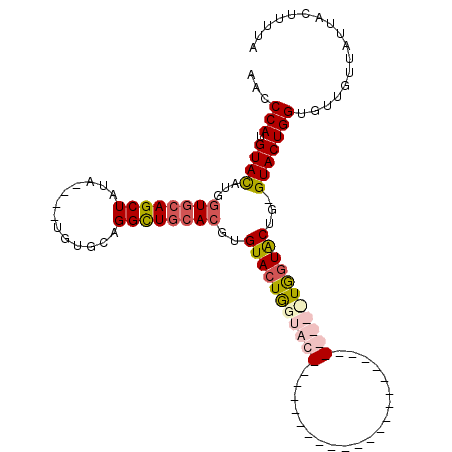
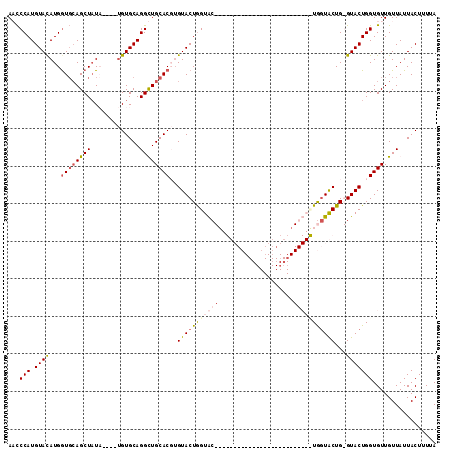
| Location | 3,764,615 – 3,764,716 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -13.49 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3764615 101 - 27905053 UAAAAGUAAUAACAACACCAGUACCCAGUACCAAUACC-------------AGUACACAGUACCAGUACACGUGCAACCUGCACAUACAUAUAGCUGCACCAUAUACAUGGGUU ......................((((((((.......(-------------(((.....(((....)))..((((.....)))).........)))).......))).))))). ( -18.74) >DroSec_CAF1 18131 110 - 1 UAAAAGUAAUAACAACACCAGUACCCAGUACCAGUACCAAUACCACUGCACAGUACACAGUACUAGUACACGUGCAACCUGCACA----UAUAGCUGGACCAUGUACAUGGAUU .................(((((.....((((.(((((.......(((....))).....))))).))))..((((.....)))).----....))))).((((....))))... ( -26.90) >DroEre_CAF1 17829 83 - 1 UAAAAGUAAUAACAACACCAGUAC-CAGCAGCA--------------------------GUACCAGUACAAGUGCAGCCUGCACG----AAUAGCUGCACCAUGUACAUGGGGU ................(((.....-..(((((.--------------------------(((....)))..((((.....)))).----....))))).((((....))))))) ( -23.60) >DroYak_CAF1 20544 83 - 1 UAAAAGUAAUAACAACACCAGUAC-CAGUACUA--------------------------GCACCAGUACCAGUGCAGCCUGCACA----AAGAGCUCCAUCAUGUACAUGGGGU ................(((((((.-...)))).--------------------------...(((((((..(((.(((((.....----.)).))).)))...)))).)))))) ( -18.10) >consensus UAAAAGUAAUAACAACACCAGUAC_CAGUACCA__________________________GUACCAGUACAAGUGCAACCUGCACA____AAUAGCUGCACCAUGUACAUGGGGU .................(((((((.((((..............................(((....)))..((((.....)))).........))))......)))).)))... (-13.49 = -14.05 + 0.56)

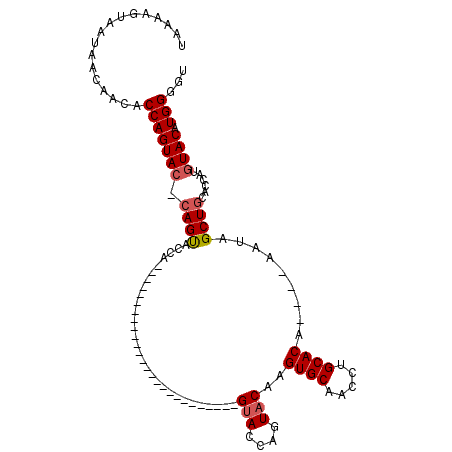
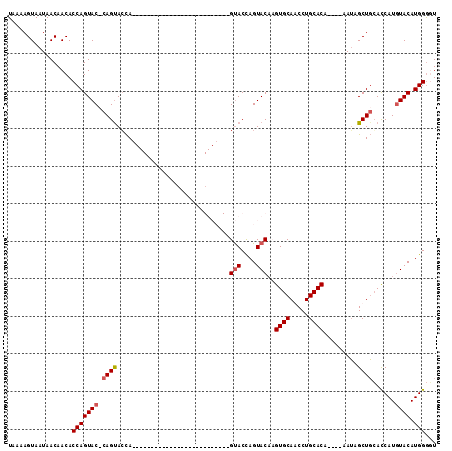
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:44 2006