
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,761,253 – 3,761,422 |
| Length | 169 |
| Max. P | 0.936246 |

| Location | 3,761,253 – 3,761,355 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -24.39 |
| Energy contribution | -24.59 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3761253 102 - 27905053 UACUCCCGUCAACAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUUC-AUUAACU-----AGAGUAACGAAUA------------C (((((((((((..................))))))))))).((((((........))))))...((.(((((.......))-))).)).-----.............------------. ( -29.87) >DroPse_CAF1 16019 102 - 1 UACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUUC-AUUAAUU-----GAAGUAACGAAUA------------U (((((((((((..(((.......)))...))))))))))).(((.(((........))).((((...(((((.......))-))).)))-----).)))........------------. ( -27.10) >DroGri_CAF1 17913 102 - 1 UACUCCCGUCAACACAAUUUGAAUAUCUAUGACGGGGGUGAGUUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUUAUACUAAUU-----AAAUUGCC-AACA------------A (((((((((((..................))))))))))).((((((........)))((((((.(((((((......)))).)))...-----..))))))-))).------------. ( -30.27) >DroEre_CAF1 14603 102 - 1 UACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUUC-AUUAACU-----AGAGUAACAUGUA------------C (((((((((((..(((.......)))...))))))))))).((((((........))))))...((.(((((.......))-))).)).-----.............------------. ( -30.90) >DroMoj_CAF1 16303 100 - 1 UACUCCCGUCAAAACAAUUUAAAUAUUUAUGACGGGGGUGAGCUACGUAACACAACGCGGCAAUGUGGAUGAUAAUUGUUCUAUUAAUC-----AAUUUG---AAUA------------U (((((((((((..................))))))))))).((..(((......)))..))....(((.(((((.......))))).))-----).....---....------------. ( -24.27) >DroAna_CAF1 13738 119 - 1 UACUCCCGUCAGCACAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUUC-AUGAACUAAACUAAAGUAACGAUAAUUGCGAUAAUCCU (((((((((((..................))))))))))).((((((........)))))).....((((((.......))-...............((((......))))....)))). ( -31.07) >consensus UACUCCCGUCAACACAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUUC_AUUAACU_____AAAGUAACGAAUA____________U (((((((((((..................))))))))))).((((((........))))))........................................................... (-24.39 = -24.59 + 0.20)



| Location | 3,761,275 – 3,761,394 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.09 |
| Mean single sequence MFE | -33.59 |
| Consensus MFE | -26.41 |
| Energy contribution | -26.05 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3761275 119 - 27905053 UCGAUCUGAUCUA-AACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAACAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUU ...(((..(((((-......(((....(((((((......(((((((((((..................))))))))))).((...))..))))))).)))....))))))))....... ( -33.47) >DroVir_CAF1 16389 120 - 1 UCGAUCUGAUCAAUCAUUAGGCCAGUAUGUUGUCAAUGAAUACUCCCGUCAAAACAAUUUAAAUAUCUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUU ........((((.((((...(((.((.((((((....(..(((((((((((..................)))))))))))..)...))))))..))..)))...)))).))))....... ( -35.37) >DroGri_CAF1 17935 120 - 1 UUGAUUUGAUCAAUUACCAGGCCAAUAUGUUGUGAAUGAAUACUCCCGUCAACACAAUUUGAAUAUCUAUGACGGGGGUGAGUUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUU ........((((.((((...(((....((((((((((...(((((((((((..................))))))))))).))).)))))))......)))...)))).))))....... ( -34.57) >DroWil_CAF1 19480 108 - 1 UCGAU------------CAGGCCAAUAUGUUGUAAAUGAAUACUCCCGUCAACAUAAUUUGAAUAUGUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUU ...((------------((..(((....(((((.......((((((((((((((((.......))))).))))))))))).....(((........)))))))).))).))))....... ( -34.50) >DroMoj_CAF1 16323 113 - 1 UCGAUCUGG-------AUAGGCCAAUAUGUUGUCAAUGAAUACUCCCGUCAAAACAAUUUAAAUAUUUAUGACGGGGGUGAGCUACGUAACACAACGCGGCAAUGUGGAUGAUAAUUGUU ......(((-------.....)))...(((..((......(((((((((((..................))))))))))).((..(((......)))..))......))..)))...... ( -29.37) >DroAna_CAF1 13777 119 - 1 UCGAUCUGAUUCA-AACCAGGCCAAUACGUUGUGAAUGAAUACUCCCGUCAGCACAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUU ..(..(((.....-...)))..)..((((((.........(((((((((((..................))))))))))).((((((........))))))))))))............. ( -34.27) >consensus UCGAUCUGAUC_A__ACCAGGCCAAUAUGUUGUGAAUGAAUACUCCCGUCAACACAAUUUGAAUAUCUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAUAAUUUUU ......................((.((((((((.......(((((((((((..................))))))))))).....(((........)))))))))))..))......... (-26.41 = -26.05 + -0.36)



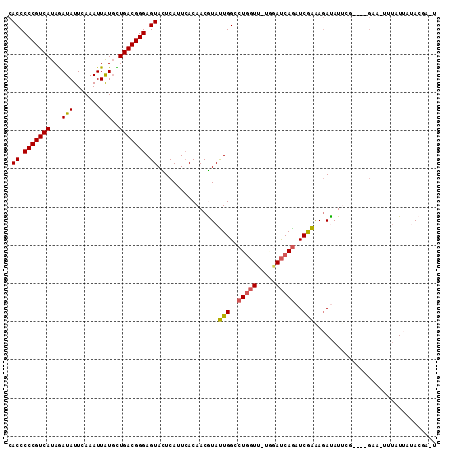
| Location | 3,761,315 – 3,761,422 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -18.07 |
| Energy contribution | -17.78 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3761315 107 + 27905053 CACCCCCGUCAUAGAUAUUCAAAUUAUGUUGACGGGAGUACUCAUUCACAACGUAUUGGCCUGGUU-UAGAUCAGAUCGAGAGACAUUCG----GCAUUUUAUUAUACAAUU .((.(((((((.(.(((.......))).)))))))).)).............((.(((..(((((.-...)))))..)))...)).....----.................. ( -21.80) >DroPse_CAF1 16081 105 + 1 CACCCCCGUCAUAGAUAUUCAAAUUAUGCUGACGGGAGUACUCAUUCACAACGUACUGGCCUGGUU-UGGAUCAGAUCGGAAGAUAUUGG----GAG-UUUAUUAUACGU-U .((.(((((((...(((.......)))..))))))).))..........((((((((((((.....-.)).))))(((....))).....----...-.......)))))-) ( -26.10) >DroGri_CAF1 17975 110 + 1 CACCCCCGUCAUAGAUAUUCAAAUUGUGUUGACGGGAGUAUUCAUUCACAACAUAUUGGCCUGGUAAUUGAUCAAAUCAAAACAUAUAUUUAUGGGUAUAUAUAGCA--AAC ....(((((((.(.(((.......))).)))))))).((((((((..........((((..((((.....))))..))))...........))))))))........--... ( -19.60) >DroMoj_CAF1 16363 100 + 1 CACCCCCGUCAUAAAUAUUUAAAUUGUUUUGACGGGAGUAUUCAUUGACAACAUAUUGGCCUAU-------CCAGAUCGAAAGAUAUGUA---GGUAAUAUGUUAUA--GUU .((.(((((((.(((((.......)))))))))))).))..........(((((((((.(((((-------....(((....)))..)))---)))))))))))...--... ( -32.30) >DroAna_CAF1 13817 104 + 1 CACCCCCGUCAUAGAUAUUCAAAUUGUGCUGACGGGAGUAUUCAUUCACAACGUAUUGGCCUGGUU-UGAAUCAGAUCGAAAGACAUUCG----CCAUUUAAU---ACGAGU .((.(((((((..(((......)))....))))))).))............((((((((((((((.-...))))).((....)).....)----)))....))---)))... ( -27.10) >DroPer_CAF1 16529 105 + 1 CACCCCCGUCAUAGAUAUUCAAAUUAUGCUGACGGGAGUACUCAUUCACAACGUACUGGCCUGGUU-UGGAUCAGAUCGGAAGAUAUUGG----GAA-UUUAUUAUACGU-U .((.(((((((...(((.......)))..))))))).))..........((((((((((((.....-.)).))))(((....))).....----...-.......)))))-) ( -26.10) >consensus CACCCCCGUCAUAGAUAUUCAAAUUAUGCUGACGGGAGUACUCAUUCACAACGUAUUGGCCUGGUU_UGGAUCAGAUCGAAAGAUAUUCG____GAA_UUUAUUAUACGA_U .((.(((((((...(((.......)))..))))))).))................(((..(((((.....)))))..)))................................ (-18.07 = -17.78 + -0.29)


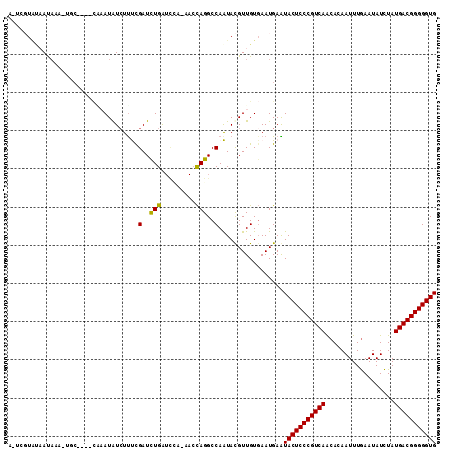
| Location | 3,761,315 – 3,761,422 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -18.08 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3761315 107 - 27905053 AAUUGUAUAAUAAAAUGC----CGAAUGUCUCUCGAUCUGAUCUA-AACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAACAUAAUUUGAAUAUCUAUGACGGGGGUG ....((((..........----.((.......))(..(((.....-...)))..).))))............(((((((((((..................))))))))))) ( -20.27) >DroPse_CAF1 16081 105 - 1 A-ACGUAUAAUAAA-CUC----CCAAUAUCUUCCGAUCUGAUCCA-AACCAGGCCAGUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUG (-((((((......-...----.....(((....)))(((.....-...)))....))))))).........(((((((((((..(((.......)))...))))))))))) ( -23.50) >DroGri_CAF1 17975 110 - 1 GUU--UGCUAUAUAUACCCAUAAAUAUAUGUUUUGAUUUGAUCAAUUACCAGGCCAAUAUGUUGUGAAUGAAUACUCCCGUCAACACAAUUUGAAUAUCUAUGACGGGGGUG (((--..(.((((((.((((((....))))..(((((...)))))......))...)))))).)..)))...(((((((((((..................))))))))))) ( -23.67) >DroMoj_CAF1 16363 100 - 1 AAC--UAUAACAUAUUACC---UACAUAUCUUUCGAUCUGG-------AUAGGCCAAUAUGUUGUCAAUGAAUACUCCCGUCAAAACAAUUUAAAUAUUUAUGACGGGGGUG ...--.((((((((((.((---((((.(((....))).)).-------.))))..)))))))))).......(((((((((((..................))))))))))) ( -27.87) >DroAna_CAF1 13817 104 - 1 ACUCGU---AUUAAAUGG----CGAAUGUCUUUCGAUCUGAUUCA-AACCAGGCCAAUACGUUGUGAAUGAAUACUCCCGUCAGCACAAUUUGAAUAUCUAUGACGGGGGUG ((.(((---((....(((----((((.....)))...(((.....-...))))))))))))..)).......(((((((((((..................))))))))))) ( -28.07) >DroPer_CAF1 16529 105 - 1 A-ACGUAUAAUAAA-UUC----CCAAUAUCUUCCGAUCUGAUCCA-AACCAGGCCAGUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUG (-((((((......-...----.....(((....)))(((.....-...)))....))))))).........(((((((((((..(((.......)))...))))))))))) ( -23.50) >consensus A_UCGUAUAAUAAA_UGC____CAAAUAUCUUUCGAUCUGAUCCA_AACCAGGCCAAUACGUUGUGAAUGAAUACUCCCGUCAACACAAUUUGAAUAUCUAUGACGGGGGUG ..................................(..(((.........)))..).................(((((((((((..................))))))))))) (-18.08 = -17.80 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:42 2006