
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,747,576 – 3,747,800 |
| Length | 224 |
| Max. P | 0.873377 |

| Location | 3,747,576 – 3,747,672 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3747576 96 - 27905053 UAA-GGCAAAAGAUAGUCUUUGGUUUUUAGGUACUAGCAAUAAUUGCAUAGGACUUGAAAAUACAUCCUAAGACUAUAUAUUAGAC-GAACUAACUUG (((-(.......((((((((.(((((((((((.((((((.....))).))).))))))))).....)).))))))))...((((..-...)))))))) ( -21.90) >DroSec_CAF1 1219 91 - 1 AAA-GGCAAAAGAUAGUCU-UGGUUUUUAGGUACUAGCAAUAAUUGCAGAGGACUUGAAAAUACAUCCUAAGACUAUUUAUUCGAC-GAA----CUUG ...-......(((((((((-((((((((((((.((.(((.....)))..)).)))))))))......)))))))))))).......-...----.... ( -21.40) >DroSim_CAF1 1215 91 - 1 AAA-CGCAAAAGAUAGUCU-UGGUUUUUAGGUACUAACAAUAAUUGCAGAGGACUUGAAAAUACAUCCUAAGACUAUUUAUUCGAC-GAA----CUUU ...-((.((.(((((((((-((((((((((((.((..((.....))...)).)))))))))......)))))))))))).))))..-...----.... ( -19.80) >DroEre_CAF1 1212 92 - 1 AAAAGGCAAAA-AUAGUCGUUGGUUUUCAAGUUU-CUGAAUAAUUGCAGAGGCUUUGAAAAUAUCUCCCAAGACUGU-UAUUCACA-GAACUAACU-- ..........(-((((((.(((((((((((((((-(((........))))))).)))))))......))))))))))-).......-.........-- ( -22.50) >DroYak_CAF1 1173 96 - 1 AAGAGGCAAAAUAUAGUCGUCGGUUUUAAAGUUU-UCCCAUAAUUGCAGAGGAUUGGAAAAUAUCUCCUAAGACUAU-AAUUCUAACUAACUAACUUG ...........(((((((...((.......((((-(((.((..((...))..)).)))))))....))...))))))-)................... ( -13.70) >consensus AAA_GGCAAAAGAUAGUCUUUGGUUUUUAGGUACUAGCAAUAAUUGCAGAGGACUUGAAAAUACAUCCUAAGACUAU_UAUUCGAC_GAACUAACUUG ............((((((((.(((((((((((.(((............))).))))))))).....)).))))))))..................... ( -9.90 = -10.90 + 1.00)



| Location | 3,747,672 – 3,747,774 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -31.77 |
| Energy contribution | -31.08 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3747672 102 + 27905053 CUCCAUGUGAUGAGUCAGAACGGAUAUCUGAGAAGCGUGCUCAUACUUGGCAGUCGCUGGGCACGAGUUACUGUGCGGUGGCUGUCCAAGAGCAGGCCAUGA ...((((.......(((((.......)))))...((.(((((....(((((((((((((.(((........))).))))))))).))))))))).)))))). ( -39.90) >DroSec_CAF1 1310 102 + 1 GUUCAUGUGAUGAGUCAGAACGGAUAUCUGAGAUGCGUGCUCAUACUUGGGAGUCGCUGGGCACGAGUUACUGUGCGGUGGCUGUCCAAGAGCAGGCCAUGA ..(((((.......(((((.......)))))...((.(((((....(((((((((((((.(((........))).)))))))).)))))))))).))))))) ( -37.70) >DroSim_CAF1 1306 102 + 1 GUUCAUGUGAUGAGUCAGAACGGAUAUCUGAGAUGCGUGCUCAUACUUGGAAGUCGCUGGGCACGAGUUACUGUGCGGUGGCUGUCCAAGAGCAGGCCAUGA ..(((((.......(((((.......)))))...((.(((((....(((((((((((((.(((........))).)))))))).)))))))))).))))))) ( -38.20) >DroEre_CAF1 1304 102 + 1 GUCCAUGUGAUGACUCAGAACGGAUAUCUGAGAAGCGUGCUCAUACUUGGGAGCCGCUGGGCACGAGUUACUGUGCGGCGGCUGUUCAAGAGCACGCCAAGA .............((((((.......))))))..((((((((....(((((((((((((.(((........))).)))))))).)))))))))))))..... ( -42.80) >DroYak_CAF1 1269 102 + 1 GUCCAUGUGAUGAGUCAGAACGGAUAUCUGAGAAGCGUGUUCAUACUUGGGAGUCGCUGGGCACGAGUUACUGUGCGGCGGCUGUUCAAGAGCAGGCCAUGA ...((((.......(((((.......)))))...((.(((((....(((((((((((((.(((........))).)))))))).)))))))))).)))))). ( -33.90) >DroAna_CAF1 1195 100 + 1 GUAAUCAUGAUGAGUCAGAACGGAUAUCUGAGUAACUUGCUUAUAUUUGGG-GUCGUUGGGCACGAGUUACUGUGCGACGGUCAU-CAAGAGCAAGUCAAGA ....((.(((..(.(((((.......))))).)..(((((((....(((((-..(((((.(((........))).)))))..).)-))))))))))))).)) ( -28.50) >consensus GUCCAUGUGAUGAGUCAGAACGGAUAUCUGAGAAGCGUGCUCAUACUUGGGAGUCGCUGGGCACGAGUUACUGUGCGGCGGCUGUCCAAGAGCAGGCCAUGA ...((((.......(((((.......)))))...((.(((((....(((((((((((((.(((........))).)))))))).)))))))))).)))))). (-31.77 = -31.08 + -0.69)

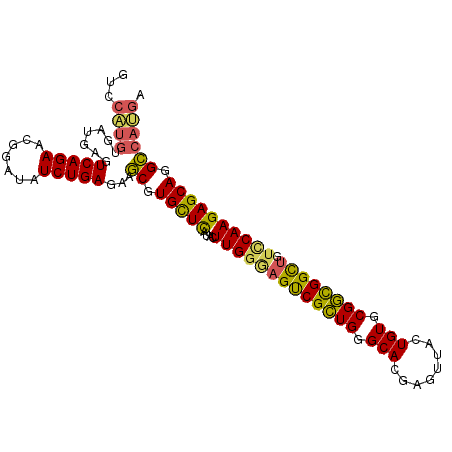
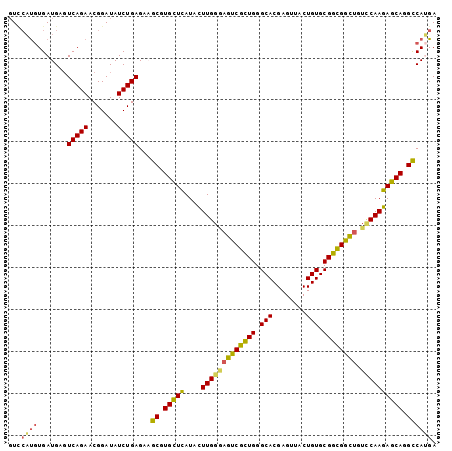
| Location | 3,747,706 – 3,747,800 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -27.22 |
| Energy contribution | -26.08 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3747706 94 + 27905053 GCGUGCUCAUACUUGGCAGUCGCUGGGCACGAGUUACUGUGCGGUGGCUGUCCAAGAGCAGGCCAUGAUUAAAACCUCGAAUACUGAAUAAGUG ((.(((((....(((((((((((((.(((........))).))))))))).))))))))).))..................((((.....)))) ( -33.70) >DroSec_CAF1 1344 94 + 1 GCGUGCUCAUACUUGGGAGUCGCUGGGCACGAGUUACUGUGCGGUGGCUGUCCAAGAGCAGGCCAUGAUUAAAACCUCGAAUACUGAAUAAGUG ((.(((((....(((((((((((((.(((........))).)))))))).)))))))))).))..................((((.....)))) ( -31.00) >DroSim_CAF1 1340 94 + 1 GCGUGCUCAUACUUGGAAGUCGCUGGGCACGAGUUACUGUGCGGUGGCUGUCCAAGAGCAGGCCAUGAUUAAAACCUCGAAUACUGAUUAAGUG ((.(((((....(((((((((((((.(((........))).)))))))).)))))))))).))..................((((.....)))) ( -31.50) >DroEre_CAF1 1338 94 + 1 GCGUGCUCAUACUUGGGAGCCGCUGGGCACGAGUUACUGUGCGGCGGCUGUUCAAGAGCACGCCAAGAUUAAAACGUCGAGUACUGAACAGUUG ((((((((....(((((((((((((.(((........))).)))))))).))))))))))))).........(((.(((.....)))...))). ( -36.60) >DroYak_CAF1 1303 94 + 1 GCGUGUUCAUACUUGGGAGUCGCUGGGCACGAGUUACUGUGCGGCGGCUGUUCAAGAGCAGGCCAUGAUUAAAGCUUCGAAUACUGAAUAAUUG ..((((((...((((((((((((((.(((........))).)))))))).)))))).(.((((..........))))))))))).......... ( -29.70) >DroAna_CAF1 1229 91 + 1 ACUUGCUUAUAUUUGGG-GUCGUUGGGCACGAGUUACUGUGCGACGGUCAU-CAAGAGCAAGUCAAGAUUAAAGAUUCCAAGCCUGAAA-GGUU ((((((((....(((((-..(((((.(((........))).)))))..).)-))))))))))).................((((.....-)))) ( -25.80) >consensus GCGUGCUCAUACUUGGGAGUCGCUGGGCACGAGUUACUGUGCGGCGGCUGUCCAAGAGCAGGCCAUGAUUAAAACCUCGAAUACUGAAUAAGUG ((.(((((....(((((((((((((.(((........))).)))))))).)))))))))).))............................... (-27.22 = -26.08 + -1.13)

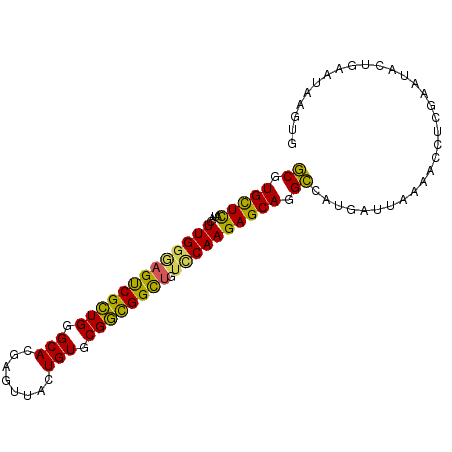

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:33 2006