
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,709,173 – 3,709,344 |
| Length | 171 |
| Max. P | 0.987759 |

| Location | 3,709,173 – 3,709,270 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -22.79 |
| Energy contribution | -24.48 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3709173 97 + 27905053 AACUUAGAAACACGGCAAAGAGUGGAAACUAUGUUGUGCAUGGAAGUGGCCAACUCCUGGCAGUUUUCUGCAUCCAGAAGAUAGCCGAGCAACAAAG .((((.....(((((((...(((....))).))))))).....))))......(((..(((.((((((((....)))))))).))))))........ ( -27.10) >DroSec_CAF1 67048 97 + 1 AACUUGGUAACACGGCAAAGAGUGGAAACUAUGUUGUGCAUGGAAGUGGCCAACUCCUGGCAGUUUCCUGCAUCCAGAAGAUAGCCGAGCAACAAAG ..((((((...((((((...(((....))).))))))(((.(((((..((((.....))))..))))))))............))))))........ ( -31.20) >DroSim_CAF1 70245 97 + 1 AACUUGGUAACACGGCAAAGAGUGGAAACUAUGUUGUGCAUGGAAGUGGCCAACUCCUGGCAGUUUCCUGCAUCCAGAAGAUAGCCGAGCAACAAAG ..((((((...((((((...(((....))).))))))(((.(((((..((((.....))))..))))))))............))))))........ ( -31.20) >DroYak_CAF1 66000 97 + 1 AACUUGGUUACACGGCAAAGAGUUUAAACUAUGCUGUGUAUAAAAGUGGCCAACUCUUCGCAGUUUCCUGCAUCCAUAAGAUAGCCAAGCAACAAAG .((((...(((((((((...(((....))).)))))))))...))))(((.........((((....))))(((.....))).)))........... ( -26.10) >consensus AACUUGGUAACACGGCAAAGAGUGGAAACUAUGUUGUGCAUGGAAGUGGCCAACUCCUGGCAGUUUCCUGCAUCCAGAAGAUAGCCGAGCAACAAAG ..((((((...((((((...(((....))).))))))(((.(((((..((((.....))))..))))))))............))))))........ (-22.79 = -24.48 + 1.69)

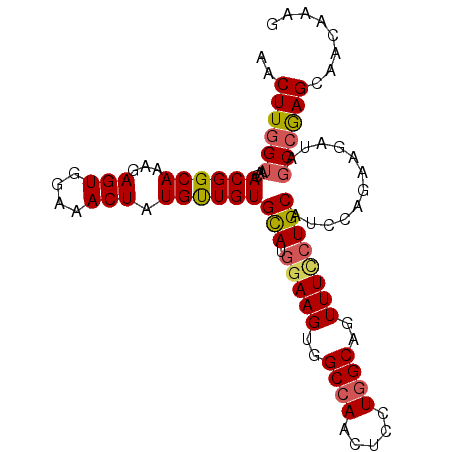
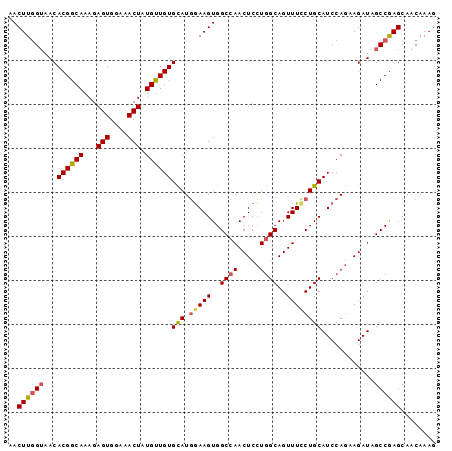
| Location | 3,709,173 – 3,709,270 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -26.71 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3709173 97 - 27905053 CUUUGUUGCUCGGCUAUCUUCUGGAUGCAGAAAACUGCCAGGAGUUGGCCACUUCCAUGCACAACAUAGUUUCCACUCUUUGCCGUGUUUCUAAGUU ...........(((((.((((((...((((....)))))))))).)))))((((....((((..((.((........)).))..))))....)))). ( -25.80) >DroSec_CAF1 67048 97 - 1 CUUUGUUGCUCGGCUAUCUUCUGGAUGCAGGAAACUGCCAGGAGUUGGCCACUUCCAUGCACAACAUAGUUUCCACUCUUUGCCGUGUUACCAAGUU ...........(((((.((((((...((((....)))))))))).)))))((((....((((..((.((........)).))..))))....)))). ( -30.10) >DroSim_CAF1 70245 97 - 1 CUUUGUUGCUCGGCUAUCUUCUGGAUGCAGGAAACUGCCAGGAGUUGGCCACUUCCAUGCACAACAUAGUUUCCACUCUUUGCCGUGUUACCAAGUU ...........(((((.((((((...((((....)))))))))).)))))((((....((((..((.((........)).))..))))....)))). ( -30.10) >DroYak_CAF1 66000 97 - 1 CUUUGUUGCUUGGCUAUCUUAUGGAUGCAGGAAACUGCGAAGAGUUGGCCACUUUUAUACACAGCAUAGUUUAAACUCUUUGCCGUGUAACCAAGUU (((.(((((.((((............((((....))))((((((((....(((..............)))...)))))))))))).))))).))).. ( -29.44) >consensus CUUUGUUGCUCGGCUAUCUUCUGGAUGCAGGAAACUGCCAGGAGUUGGCCACUUCCAUGCACAACAUAGUUUCCACUCUUUGCCGUGUUACCAAGUU ...........(((((.((((((...((((....)))))))))).)))))((((....((((..((.((........)).))..))))....)))). (-26.71 = -27.02 + 0.31)

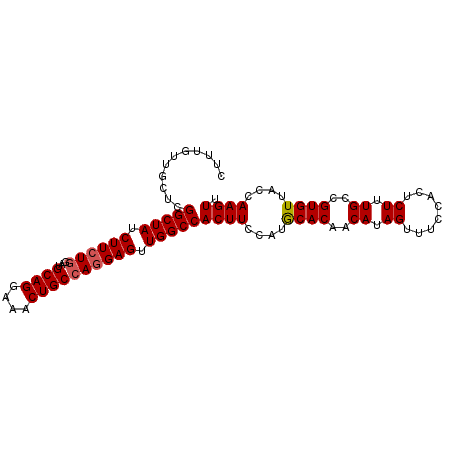
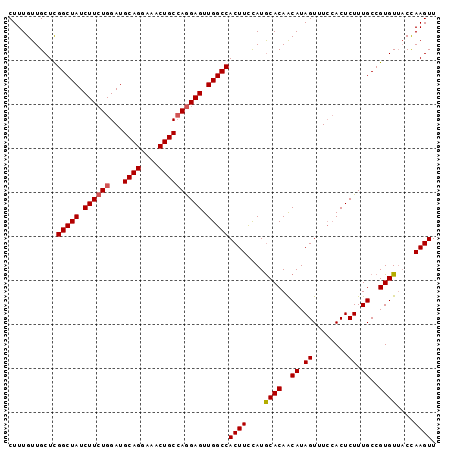
| Location | 3,709,207 – 3,709,304 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.63 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -10.86 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.28 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3709207 97 - 27905053 UUCCGCUG-GAUUGUGUAGAACGCGAGGCGGCUG-CCUUUGUUGCUCGGC-UAUCUUCUGGAUGCAGAAAACUGCCAGGA---------------GUUGGCCACUUCCAUGCACA ....((((-(((((((.....)))))((((((..-.....)))))).(((-((.((((((...((((....)))))))))---------------).)))))...)))).))... ( -36.00) >DroPse_CAF1 69776 105 - 1 UGUGGCUGUGACUGUGGGGC---UGCUGCUGCUGUCUUUUGUUCCUCGUCUUAUCUUAUGGGAGCAGGAAGCUGUGGGGAUAAGGAUGAGGAUGAGUCGUCC-------UGCCUG ...(((.((((((..(((((---.((....)).)))))....(((((((((((((((......((((....)))).))))))).))))))))..))))).).-------.))).. ( -37.60) >DroSec_CAF1 67082 97 - 1 UUCCGCUG-GAUUGUGUAGAAGGCGAGGCGCCUG-CCUUUGUUGCUCGGC-UAUCUUCUGGAUGCAGGAAACUGCCAGGA---------------GUUGGCCACUUCCAUGCACA ....((((-((..(.(((((((((.((....)))-)))))..))).)(((-((.((((((...((((....)))))))))---------------).)))))...)))).))... ( -41.70) >DroSim_CAF1 70279 97 - 1 UUCCGCUG-GAUUGUGUAGAAGGCGAGGCGGCUG-CCUUUGUUGCUCGGC-UAUCUUCUGGAUGCAGGAAACUGCCAGGA---------------GUUGGCCACUUCCAUGCACA ....((((-((..(.(((((((((.((....)))-)))))..))).)(((-((.((((((...((((....)))))))))---------------).)))))...)))).))... ( -40.20) >DroYak_CAF1 66034 97 - 1 UUCUACUG-GAUUGUAUAGAAGGCGAGGCGGUUG-CCUUUGUUGCUUGGC-UAUCUUAUGGAUGCAGGAAACUGCGAAGA---------------GUUGGCCACUUUUAUACACA ........-...(((((((((((((((((....)-))))....)).((((-((.(((.....(((((....)))))..))---------------).)))))))))))))))).. ( -34.60) >DroPer_CAF1 75626 105 - 1 UGUGGCUGUGGCUGUGGGGC---UGCUGCUGCUGUCUUUUGUUCCUCGUCUUAUCUUAUGGGAGCAGGAAACUGUGGGGAUAAGGAUGAGGAUGAGUCGUCC-------UGCCUG .(..((.(..(((....)))---..).))..).(.(((....(((((((((((((((......((((....)))).))))))).)))))))).))).)....-------...... ( -40.30) >consensus UUCCGCUG_GAUUGUGUAGAAGGCGAGGCGGCUG_CCUUUGUUGCUCGGC_UAUCUUAUGGAUGCAGGAAACUGCCAGGA_______________GUUGGCCACUUCCAUGCACA .............((((.((((.(((.(((((........))))))))...............((((....))))............................)))).))))... (-10.86 = -12.28 + 1.42)
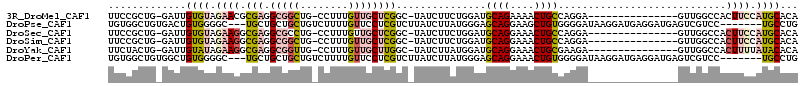
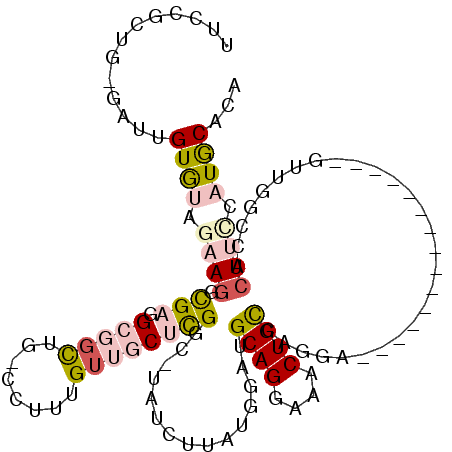

| Location | 3,709,232 – 3,709,344 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -31.41 |
| Energy contribution | -30.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3709232 112 + 27905053 GCAGUUUUCUGCAUCCAGAAGAUAGCCGAGCAACAAAGGCAGCCGCCUCGCGUUCUACACAAUCCAGCGGAAGACAUUGCGGAUGGUUAUAGAUGGAGGAAACAAUAGUCCG ((((....)))).((((....((((((((((.....((((....))))...))))......((((.((((......))))))))))))))...))))(((........))). ( -33.20) >DroSec_CAF1 67107 112 + 1 GCAGUUUCCUGCAUCCAGAAGAUAGCCGAGCAACAAAGGCAGGCGCCUCGCCUUCUACACAAUCCAGCGGAAGACAUUGCGGAUGGUUAUAGAUGGAGGAAACAAUAUUGUG ...(((((((.((((......(((((((.((((...((((....)))).(.((((..(........)..)))).).))))...))))))).)))).)))))))......... ( -35.10) >DroSim_CAF1 70304 112 + 1 GCAGUUUCCUGCAUCCAGAAGAUAGCCGAGCAACAAAGGCAGCCGCCUCGCCUUCUACACAAUCCAGCGGAAGACAUUGCGGAUGGUUAUAGAUGGAGGAAACAAUAGUGCG ((((((((((.((((......(((((((.((((...((((....)))).(.((((..(........)..)))).).))))...))))))).)))).))))))).....))). ( -35.90) >DroYak_CAF1 66059 112 + 1 GCAGUUUCCUGCAUCCAUAAGAUAGCCAAGCAACAAAGGCAACCGCCUCGCCUUCUAUACAAUCCAGUAGAAGACAUUGCGGAUGGUUAUAGAUGUGCGAAACAAUAGGGCG ...(((((.((((((......(((((((.((((...((((....)))).(.(((((((........))))))).).))))...))))))).)))))).)))))......... ( -32.70) >consensus GCAGUUUCCUGCAUCCAGAAGAUAGCCGAGCAACAAAGGCAGCCGCCUCGCCUUCUACACAAUCCAGCGGAAGACAUUGCGGAUGGUUAUAGAUGGAGGAAACAAUAGUGCG ...(((((((.((((......(((((((.((((...((((....)))).(.(((((.(........).))))).).))))...))))))).)))).)))))))......... (-31.41 = -30.97 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:15 2006