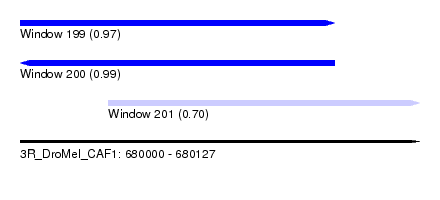
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 680,000 – 680,127 |
| Length | 127 |
| Max. P | 0.989132 |

| Location | 680,000 – 680,100 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -46.08 |
| Consensus MFE | -25.74 |
| Energy contribution | -27.55 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 680000 100 + 27905053 CUUGC--GU------GGGCAUAACCCUUG-CCCGCCU---GGGCAAAUCCCGGACGUUGUCGAUGUGAGCGUGU-UACCGACGGCAAAAGUCGUUGCAACGCCCACUACCUGG .....--((------(((((.......))-)))))((---(((.....)))))...........(((.((((..-...(((((((....)))))))..)))).)))....... ( -35.80) >DroPse_CAF1 100719 99 + 1 CUUGCC-GU------GGGUUUAACCCUUU-CCGGGGC---AAGUAAAUCCCUUCGG--CUUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGG (..(((-(.------(((((((.((((..-..)))).---...)))))))...)))--)..).(((((((((..((.(((((.......))))).)).)))))))))-..... ( -48.00) >DroEre_CAF1 96278 100 + 1 CUUGU--GU------GGGCAUAUCCCUUG-CCCGACU---GGGCAAAUCCCGCACGUUGUCGAUGUGGGCGUGU-UGCCGUCGGCAAAAGUCGGUGAUACGCCCACUACCUGG ...((--((------(((.((.....(((-(((....---))))))))))))))).........((((((((((-..(((.(.......).)))..))))))))))....... ( -48.80) >DroYak_CAF1 109311 106 + 1 CUUGU--GUAGGUAUGGGCAUAACCCUUG-CUUGACU---GGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU-UGCCGCCGGCAAAAGUCGGUGCAACGCCCACUACCUGG .....--.(((((((((((....(((...-(((((((---(((.....))))......))))))..)))...((-(((.((((((....)))))))))))))))).)))))). ( -46.50) >DroAna_CAF1 99014 106 + 1 CUUGCCGGC------GGGUUUAACCCUUUGACCCUUUGACGGGUAAAUCCCGGACGUGGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGG ....((((.------(((.....))).((((((......((((.....)))).....))))))(((((((((..-.((((((.......))))))...)))))))))..)))) ( -49.40) >DroPer_CAF1 101706 99 + 1 CUUGCC-GU------GGGUUUAACCCUUU-CCGGGGC---AUGUAAAUCCCUUCGG--CUUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGG (..(((-(.------(((((((.((((..-..)))).---...)))))))...)))--)..).(((((((((..((.(((((.......))))).)).)))))))))-..... ( -48.00) >consensus CUUGC__GU______GGGCAUAACCCUUG_CCCGACU___GGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU_UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGG ...............(((.....)))..............(((.....)))............(((((((((...(((((((.......)))))))..)))))))))...... (-25.74 = -27.55 + 1.81)

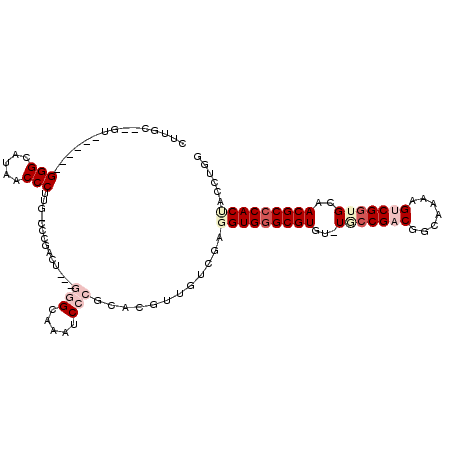
| Location | 680,000 – 680,100 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -45.07 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 680000 100 - 27905053 CCAGGUAGUGGGCGUUGCAACGACUUUUGCCGUCGGUA-ACACGCUCACAUCGACAACGUCCGGGAUUUGCCC---AGGCGGG-CAAGGGUUAUGCCC------AC--GCAAG .......((((((((......((((((((((((((((.-..........))))))..((((.(((.....)))---.))))))-))))))))))))))------))--..... ( -42.80) >DroPse_CAF1 100719 99 - 1 CCAGG-GGUGGGCGUUGCACCGACUUUUGCCGUCGGAACGCACGCCCACCUCAAG--CCGAAGGGAUUUACUU---GCCCCGG-AAAGGGUUAAACCC------AC-GGCAAG ...((-(((((((((.((.(((((.......)))))...)))))))))))))..(--(((..(((.((((...---((((...-...)))))))))))------.)-)))... ( -49.80) >DroEre_CAF1 96278 100 - 1 CCAGGUAGUGGGCGUAUCACCGACUUUUGCCGACGGCA-ACACGCCCACAUCGACAACGUGCGGGAUUUGCCC---AGUCGGG-CAAGGGAUAUGCCC------AC--ACAAG ....((.(((((((((((.((.....((((((((((((-(....(((.((.((....)))).)))..))))).---.))).))-))))))))))))))------))--))... ( -45.00) >DroYak_CAF1 109311 106 - 1 CCAGGUAGUGGGCGUUGCACCGACUUUUGCCGGCGGCA-ACACGCCCACCUCGACAACGUGCGGGAUUUGCCC---AGUCAAG-CAAGGGUUAUGCCCAUACCUAC--ACAAG ..(((((.(((((((......(((((((((.(((((((-(....(((.(..((....)).).)))..))))).---.)))..)-))))))))))))))))))))..--..... ( -39.00) >DroAna_CAF1 99014 106 - 1 CCAGGUAGUGGGCGUUGCACCGACUUUUGCCGUCGGCA-ACACGCCCACCUCGACCACGUCCGGGAUUUACCCGUCAAAGGGUCAAAGGGUUAAACCC------GCCGGCAAG ...(((.((((((((....(((((.......)))))..-..))))))))....)))..(.((((..(((((((......)))).)))(((.....)))------.)))))... ( -44.00) >DroPer_CAF1 101706 99 - 1 CCAGG-GGUGGGCGUUGCACCGACUUUUGCCGUCGGAACGCACGCCCACCUCAAG--CCGAAGGGAUUUACAU---GCCCCGG-AAAGGGUUAAACCC------AC-GGCAAG ...((-(((((((((.((.(((((.......)))))...)))))))))))))..(--(((..(((.((((...---((((...-...)))))))))))------.)-)))... ( -49.80) >consensus CCAGGUAGUGGGCGUUGCACCGACUUUUGCCGUCGGCA_ACACGCCCACCUCGACAACGUACGGGAUUUACCC___AGCCGGG_AAAGGGUUAAACCC______AC__GCAAG .......((((((((....(((((.......))))).....)))))))).............(((....((((..............))))....)))............... (-25.04 = -25.24 + 0.20)
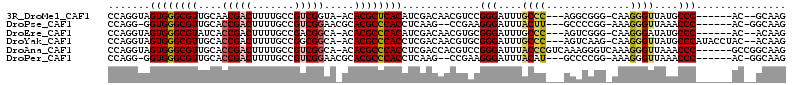
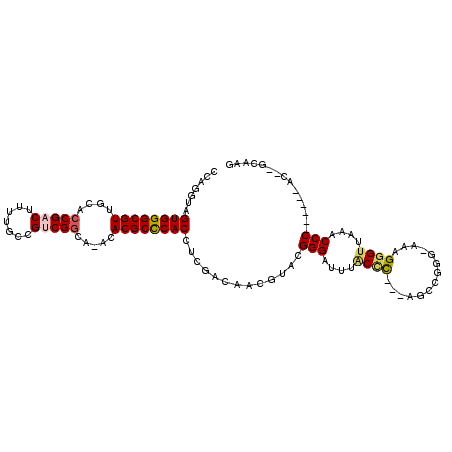
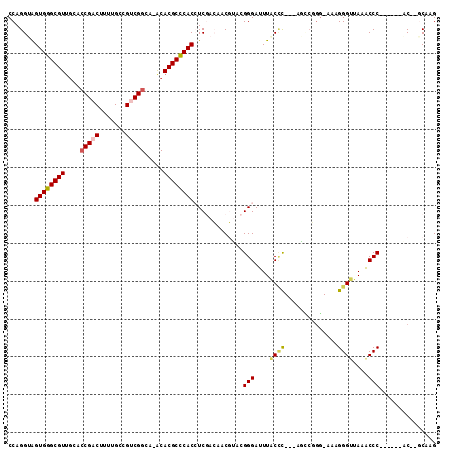
| Location | 680,028 – 680,127 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -27.43 |
| Energy contribution | -29.23 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 680028 99 + 27905053 GGGCAAAUCCCGGACGUUGUCGAUGUGAGCGUGU-UACCGACGGCAAAAGUCGUUGCAACGCCCACUACCUGGCUUAACCCGCCACUG---GCCGG-GCGGGUC (((.....))).......((((..(((.((((..-...(((((((....)))))))..)))).)))....))))...(((((((....---....)-)))))). ( -36.30) >DroPse_CAF1 100748 100 + 1 AAGUAAAUCCCUUCGG--CUUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGGCUUAACCCGCCACCAUCCUACGC-GCGGGCC ..............((--((.(.(((((((((..((.(((((.......))))).)).)))))))))-.).))))...((((((..........).-))))).. ( -42.70) >DroEre_CAF1 96306 102 + 1 GGGCAAAUCCCGCACGUUGUCGAUGUGGGCGUGU-UGCCGUCGGCAAAAGUCGGUGAUACGCCCACUACCUGGCUUAACCCGCCACUGGCCAUCGG-UCGGGCC (((.....)))((...........((((((((((-..(((.(.......).)))..))))))))))....((((.......))))((((((...))-)))))). ( -44.80) >DroYak_CAF1 109345 93 + 1 GGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU-UGCCGCCGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCG---------GUCGG-UCGGGCC ((((....(((((.((....))..)))))...((-(((.((((((....))))))))))))))).......((((..(((..---------...))-)..)))) ( -43.40) >DroAna_CAF1 99048 100 + 1 GGGUAAAUCCCGGACGUGGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCGCCACCG---GACGCUGCGAGCC (((.....)))((.((..(.((.(((((((((..-.((((((.......))))))...))))))))).((((((.......))))..)---).)))..))..)) ( -46.20) >DroPer_CAF1 101735 100 + 1 AUGUAAAUCCCUUCGG--CUUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGGCUUAACCCGCCACCAUCCUACGC-GCGGGCC ..............((--((.(.(((((((((..((.(((((.......))))).)).)))))))))-.).))))...((((((..........).-))))).. ( -42.70) >consensus GGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU_UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCGCCACCG___GACGC_GCGGGCC ........(((............(((((((((...(((((((.......)))))))..)))))))))...((((.......))))..............))).. (-27.43 = -29.23 + 1.81)

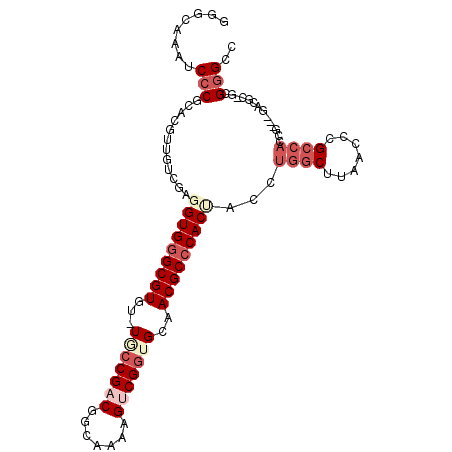
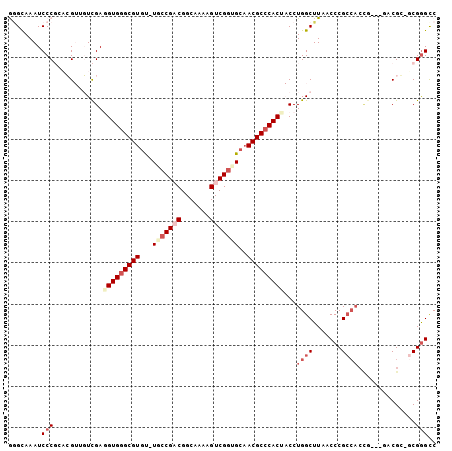
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:47 2006