
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,667,148 – 3,667,359 |
| Length | 211 |
| Max. P | 0.880373 |

| Location | 3,667,148 – 3,667,268 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -28.55 |
| Energy contribution | -31.05 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
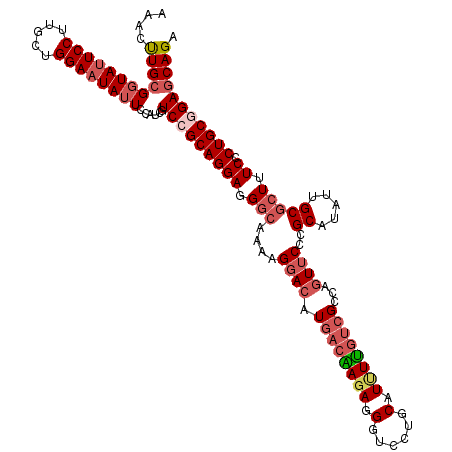
>3R_DroMel_CAF1 3667148 120 + 27905053 AAACUUGCGGUAUUCCUUGCUGGAAUAUUCCAUCGUCCGCAGGAGGGCAAAAGGACAUGACGAGAGGAUCCUGCAUUUCGUCGCCAAUUCCCGCAUAUUGCGCUUUCACUGCCGAGCAGA ....((((((((((((.....))))))..))....((.((((((((((....((....(((((((.(......).))))))).)).......((.....)))))))).)))).)))))). ( -37.90) >DroSec_CAF1 25942 120 + 1 AAACUUGCGGUAUUCCUUGCUGGAAUAUUCCAUCAUCCGCAGGAUGGCAAAAGGACAUGACAAGAGGGUCCUGCAUCUUGUCGCUAGUUCUCGCAUAUUGCGCUUUCCCUGCGGAGCAGA ....((((((((((((.....))))))..))....((((((((..(((...(((((.((((((((..(.....).))))))))...))))).((.....)))))...)))))))))))). ( -45.50) >DroSim_CAF1 28195 120 + 1 AAACUUGCGGUAUUCCUUGCUGGAAUAUUCCAUCGUCCGCAGGAUGGCAAAAGGACAUGACAAGAGGGUCCUGCAUUUUGUCGCUAGUUCUCGCAUAUUGCGCUUUCCCUGCGGACCAGA .....((..(((((((.....)))))))..))..(((((((((..(((...(((((.((((((((..(.....).))))))))...))))).((.....)))))...))))))))).... ( -43.50) >DroEre_CAF1 25403 116 + 1 AAAACUGCGCUACUCCUGGUUGGAAUAUUUCUGCGUCAGCAGGAGGCCAAAAGGACCUGAGGAGAGGGUCCUGCAUGUU---GACAGUGCCCGCAUAUUGCACU-UCCCUGCAGAGCAGA ....((((.....((((..((((....(((((((....))))))).)))).)))).((((((.(((((..((((.....---).)))..)))((.....))...-))))).))).)))). ( -44.00) >consensus AAACUUGCGGUAUUCCUUGCUGGAAUAUUCCAUCGUCCGCAGGAGGGCAAAAGGACAUGACAAGAGGGUCCUGCAUUUUGUCGCCAGUUCCCGCAUAUUGCGCUUUCCCUGCGGAGCAGA ....((((((((((((.....))))))))......(((((((((.(((....((((.((((((((.(......).))))))))...))))..((.....))))).)).))))))))))). (-28.55 = -31.05 + 2.50)

| Location | 3,667,268 – 3,667,359 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.16 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3667268 91 - 27905053 CCAGCACGGCGGGUUUCCACUUGCAAAA---AAAUAAAACCGUAAAACCGGGACACAUCAGUAUAAAACAGCAGAGAAAGACGUCGCUGGCCAU (((((..((((..((((...((((....---........(((......))).........((.....)).)))).))))..))))))))).... ( -21.80) >DroSec_CAF1 26062 92 - 1 CCAGAACGGCGGGUUUCCACUUGCAAAAA--AAAAGGAACCGUAAAACCGGGACACAUCAGUAUAAAACAGCAGAGACCAACGUCGCUGGCCAU ((((..(((((((((((....(((.....--.....(..(((......)))..)......((.....)).)))))))))..))))))))).... ( -22.10) >DroSim_CAF1 28315 91 - 1 CCAGCACAGCGAGUUUCCACUUGCAAAA---AAAUGGAACCGUAAAACCGGGACACAUCAGUAUAAAACAGCAGAGACCAACGUCGCUGGCCAU (((((((.((((((....))))))....---..((((..(((......)))..).)))..)).............(((....)))))))).... ( -21.70) >DroEre_CAF1 25519 88 - 1 CCAUCACG---GGUUUCCACUUGCAAAA---AAGUGGAACCGUAAAACUGGGACACACCAGCACAAACCAGCAGAGGCCGAUGUCGCUGGACAU .....(((---(...(((((((......---))))))).))))....((((......))))......(((((...(((....)))))))).... ( -29.00) >DroYak_CAF1 17493 91 - 1 CCAUCACA---GGUUUCAACUUGCAAAAAAAAAAUGGAACCGAAUAACUGGGACAAACCAGUAUAAAAUAGCAGAAGCCGAUGUCGCUGGCCAU (((.((((---((((((....(((..........((....))....(((((......)))))........)))))))))..))).).))).... ( -18.90) >consensus CCAGCACGGCGGGUUUCCACUUGCAAAA___AAAUGGAACCGUAAAACCGGGACACAUCAGUAUAAAACAGCAGAGACCGACGUCGCUGGCCAU ((((((((...((((((....(((...............(((......)))(......)...........)))))))))..))).))))).... (-15.10 = -15.58 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:52 2006