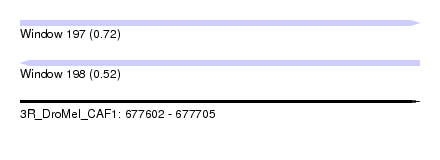
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 677,602 – 677,705 |
| Length | 103 |
| Max. P | 0.720570 |

| Location | 677,602 – 677,705 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.12 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -15.29 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 677602 103 + 27905053 AUAUACAUA-----CAGCAUAGUGUUAUACCAUAUUAUCACAAAGCAAAUGUUUUGUAUGAAAAUAAAUUUAUACAUUUUUGCCCUAUGCAGGCGAAGUAUCACUUCA .........-----..(((((((((.(((......))).)))..(((((.....((((((((......))))))))..))))).))))))....((((.....)))). ( -21.60) >DroSec_CAF1 96908 108 + 1 AUAUGCCUAUGGACCAGCCUUAUGUUAUACCAUAUUAUCACAAAACAAAUGUUUUGUAUGAAAAUAAAUUUAUACAUUUUCGCCCCAUCCAGGCGAAGUAUCACUUCA ....((((((((....((...((((.......((((.(((((((((....)))))))..)).)))).......))))....)).))))..))))((((.....)))). ( -22.24) >DroSim_CAF1 95316 108 + 1 AUAUGCCUAUGGACCAGCAUAAUGUUAUACCAUAUUAUCACAAAACAAAUGUUUUGUAUGAAAAUAGAUUUAUACAUUUUCUCCCCAUCCAGGCGAAGUAUCACUUCA ....((((((((...((...(((((.......((((.(((((((((....)))))))..)).)))).......)))))..))..))))..))))((((.....)))). ( -23.64) >consensus AUAUGCCUAUGGACCAGCAUAAUGUUAUACCAUAUUAUCACAAAACAAAUGUUUUGUAUGAAAAUAAAUUUAUACAUUUUCGCCCCAUCCAGGCGAAGUAUCACUUCA ..((((...............((((.......((((.(((((((((....)))))))..)).)))).......)))).((((((.......))))))))))....... (-15.29 = -14.74 + -0.55)

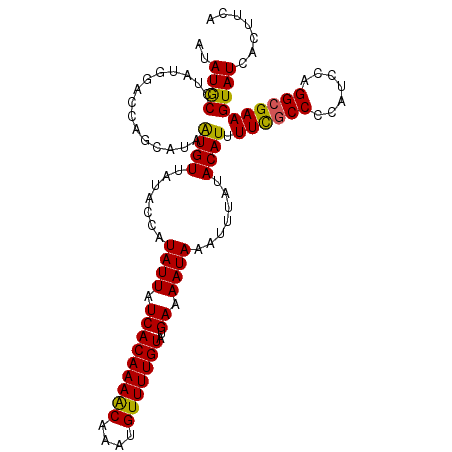
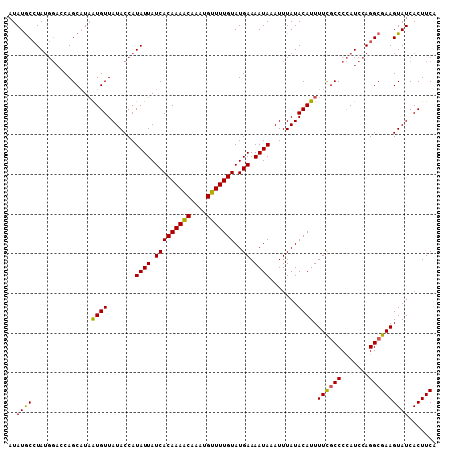
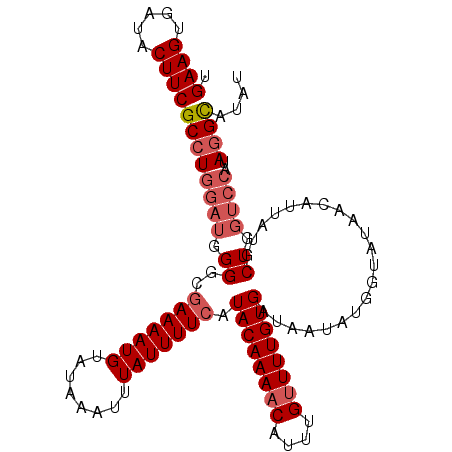
| Location | 677,602 – 677,705 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.12 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -18.19 |
| Energy contribution | -20.30 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 677602 103 - 27905053 UGAAGUGAUACUUCGCCUGCAUAGGGCAAAAAUGUAUAAAUUUAUUUUCAUACAAAACAUUUGCUUUGUGAUAAUAUGGUAUAACACUAUGCUG-----UAUGUAUAU .((((.....)))).....(((((((((((..(((((.((......)).))))).....)))))))))))...((((((((((....)))))))-----)))...... ( -23.10) >DroSec_CAF1 96908 108 - 1 UGAAGUGAUACUUCGCCUGGAUGGGGCGAAAAUGUAUAAAUUUAUUUUCAUACAAAACAUUUGUUUUGUGAUAAUAUGGUAUAACAUAAGGCUGGUCCAUAGGCAUAU .((((.....))))((((((((.((.((((((((........))))))).((((((((....))))))))....((((......)))).).)).))))..)))).... ( -33.30) >DroSim_CAF1 95316 108 - 1 UGAAGUGAUACUUCGCCUGGAUGGGGAGAAAAUGUAUAAAUCUAUUUUCAUACAAAACAUUUGUUUUGUGAUAAUAUGGUAUAACAUUAUGCUGGUCCAUAGGCAUAU .((((.....))))((((((((.((.(...(((((.((.(((((((.((..(((((((....))))))))).)))).))).))))))).).)).))))..)))).... ( -29.00) >consensus UGAAGUGAUACUUCGCCUGGAUGGGGCGAAAAUGUAUAAAUUUAUUUUCAUACAAAACAUUUGUUUUGUGAUAAUAUGGUAUAACAUUAUGCUGGUCCAUAGGCAUAU .((((.....))))((((((((.((..(((((((........))))))).((((((((....)))))))).....................)).))))..)))).... (-18.19 = -20.30 + 2.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:43 2006