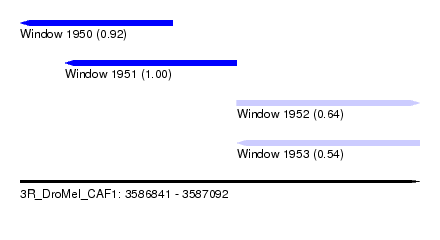
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,586,841 – 3,587,092 |
| Length | 251 |
| Max. P | 0.999659 |

| Location | 3,586,841 – 3,586,937 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.01 |
| Mean single sequence MFE | -16.83 |
| Consensus MFE | -13.84 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3586841 96 - 27905053 AUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUUUUAAUAAGAAAUUUCACAGCUUAUUUAAUAAGAACGCGUCUUU ..............(((((((((((((......(((((....)))))....((.....))))))))......(((((...)))))))))))).... ( -15.30) >DroSec_CAF1 37374 96 - 1 AUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUCUUAAUAAGAAGAUUCACAGCUUAUUCAAUAAGAACGCGUCUUU ..............(((((((......(((((((((((....)))).............)))))))......(((((...)))))))))))).... ( -17.91) >DroSim_CAF1 38176 96 - 1 AUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUCUUAAUAAGAAGAUUCACAGCUUAUUCAAUAAGAACGCGUCUUU ..............(((((((......(((((((((((....)))).............)))))))......(((((...)))))))))))).... ( -17.91) >DroEre_CAF1 42977 94 - 1 AUCCCAACUACUUUGCGUGUUGACUUCAUCUUCUGGGAUAUUUCCCACGAAUCUUAAUAAUAAAUUUCACAGCUUAUUAA--AAGAACGCGUCUUU ..............(((((((.........((((((((....))))).)))..(((((((.............)))))))--...))))))).... ( -16.42) >DroYak_CAF1 60617 94 - 1 AUCCCAACUACUUUGCGUGUUGACUUCAUCUUCUGGGAUAUUUCCCACAAGUCUUAAUAAUAAAUUUCACAGCUUGUUAA--CAGAACGCGUCUUU ..............((((.(((............((((....))))((((((...................))))))...--))).))))...... ( -16.61) >consensus AUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUCUUAAUAAGAAAUUUCACAGCUUAUUAAAUAAGAACGCGUCUUU ..............(((((((............(((((....)))))........((((((...........)))))).......))))))).... (-13.84 = -14.08 + 0.24)

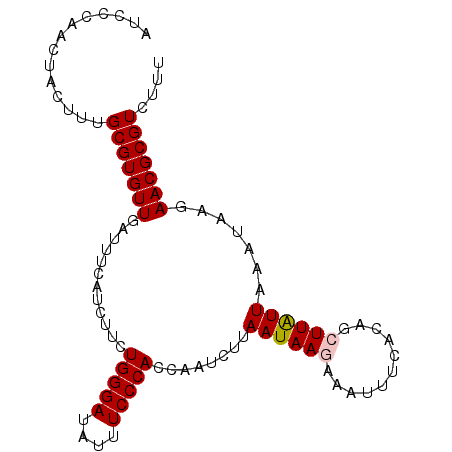
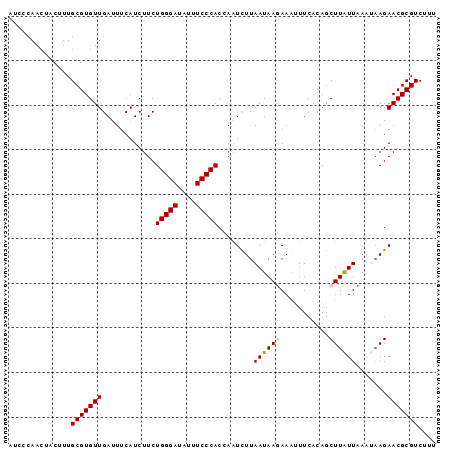
| Location | 3,586,869 – 3,586,977 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.91 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3586869 108 - 27905053 UUGUAGAACGGUGAAAGGCAGCAGGCAGGUGGCCAAAAAAAUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUUUUAAUAAGAAAUUUC ....((((.(((((((..(((((.((((((((...............)))).)))).))))).)))))))))))((((....))))...................... ( -30.46) >DroSec_CAF1 37402 108 - 1 UUGUAGAACGGUGAAAGGCAGCAGGCAGGUGGCCAAAAAAAUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUCUUAAUAAGAAGAUUC ....((((.(((((((..(((((.((((((((...............)))).)))).))))).)))))))))))((((....))))...((((((......)))))). ( -32.96) >DroSim_CAF1 38204 108 - 1 UUGUAGAACGGUGAAAGGCAGCAGGCAGGUGGCCAAAAAAAUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUCUUAAUAAGAAGAUUC ....((((.(((((((..(((((.((((((((...............)))).)))).))))).)))))))))))((((....))))...((((((......)))))). ( -32.96) >DroEre_CAF1 43003 108 - 1 UUGUAGAACGGUGAAAGGCAGCAGGCAGGUGGUCAAGAAAAUCCCAACUACUUUGCGUGUUGACUUCAUCUUCUGGGAUAUUUCCCACGAAUCUUAAUAAUAAAUUUC ((((((((.((((((...(((((.(((((((((...(.....)...))))).)))).)))))..))))))))))((((....)))))))).................. ( -31.80) >DroYak_CAF1 60643 108 - 1 UUGUAGAACGGUGAAAGGCAGAAGGCAGGUGGCCCAAAAAAUCCCAACUACUUUGCGUGUUGACUUCAUCUUCUGGGAUAUUUCCCACAAGUCUUAAUAAUAAAUUUC ((((((((.((((((...(((.(.((((((((...............)))).)))).).)))..))))))))))((((....)))))))).................. ( -23.96) >consensus UUGUAGAACGGUGAAAGGCAGCAGGCAGGUGGCCAAAAAAAUCCCAACUACUUUGCGUGUUGAUUUCAUCUUCUGGGAUAUUUCCCACCAAUCUUAAUAAGAAAUUUC ....((((.((((((...(((((.((((((((...............)))).)))).)))))..))))))))))((((....))))...................... (-27.90 = -28.10 + 0.20)

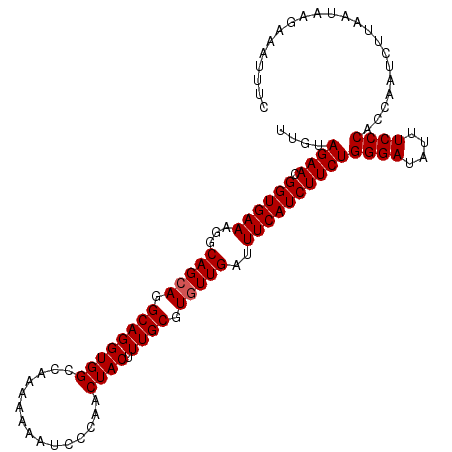
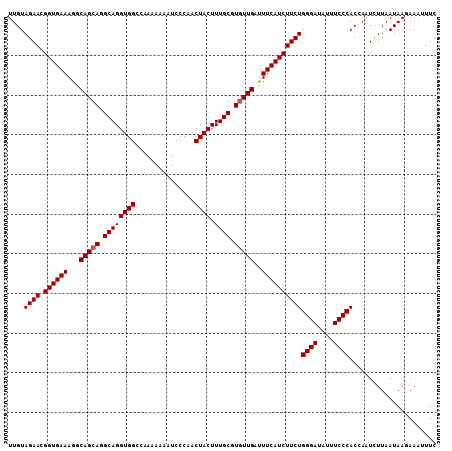
| Location | 3,586,977 – 3,587,092 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -18.76 |
| Energy contribution | -20.87 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3586977 115 + 27905053 AAUGUUGCGGCACAAGGAAGUAAGUACCGAGCUAUAAAUAUGUGAUUCACUCAGAGCUGACUCAUUAAUUGAGACCGGUACUUAUCCGU---GUUAAGCGCAAUAGCUUAACAGAA--UU ......((((.........((((((((((((((........(((...)))....))))..((((.....))))..))))))))))))))---(((((((......)))))))....--.. ( -33.40) >DroSec_CAF1 37510 115 + 1 AAUGUUGCGGCCCAAGGAAGUAAGUACCGAGCUAUAAAUAUGUGAUUCACUCAGAGCUGACUCAUUAAUUGAGACCGGUACUUAUCCGU---GUUAAGCGCAAUAGCUUAGCAGAA--UU ......((((.........((((((((((((((........(((...)))....))))..((((.....))))..))))))))))))))---(((((((......)))))))....--.. ( -33.40) >DroSim_CAF1 38312 115 + 1 AAUGUUGCGGCCCAAGGAAGUAAGUACCGCGCUAUAAAUAUGUGAUUCACUCAGAGCUGACUCAUUAAUUGAGACCGGUACUUAUCCGU---GUUAAGCGCAAUAGCUUAGCAGAA--UU ......((((.........((((((((((.(((........(((...)))....)))...((((.....))))..))))))))))))))---(((((((......)))))))....--.. ( -31.50) >DroEre_CAF1 43111 115 + 1 AAUGUUGGGGCCCAAGGAAGUAAGUACCGAGCUAUAAACGUGUGAUUCACUCAGAGCUGACUCAUUAAUUGAGACCGGUACUUAUACUG---GUUAAGCGCAAUAGCUUAACAGAA--AU ..(.((((...)))).)..((((((((((((((......(((.....)))....))))..((((.....))))..)))))))))).(((---.((((((......)))))))))..--.. ( -34.60) >DroYak_CAF1 60751 115 + 1 AAUGUUGAGGCCCAAGGAAGUAAGUACCCAGCUAUAAAUAUGUGAUUCACUCCGAGCUGACUCAUUAAUUGAGACCGGUACUUAGACUC---GUUAAGCGCAACAGCUUAACAGAA--AU ......(((.((...))...(((((((((((((........(((...)))....))))).((((.....))))...))))))))..)))---(((((((......)))))))....--.. ( -32.00) >DroAna_CAF1 34402 111 + 1 ACAGUUCAAGCCCAACAAAGUUAGUACA----UCUUAAAACCUGAAUCACCCAGCACUGACUCAUGAUCAGUAGCCGGCAGCUAUCGACGUCAUUAAGGGAA-----UUAACAGAGAGUU ...(((....(((.....(((((((...----.........(((.......))).))))))).((((((.(((((.....))))).))..))))...)))..-----..)))........ ( -17.72) >consensus AAUGUUGCGGCCCAAGGAAGUAAGUACCGAGCUAUAAAUAUGUGAUUCACUCAGAGCUGACUCAUUAAUUGAGACCGGUACUUAUCCGU___GUUAAGCGCAAUAGCUUAACAGAA__UU ...................((((((((((..((........((((.(((........))).))))......))..)))))))))).......(((((((......)))))))........ (-18.76 = -20.87 + 2.11)

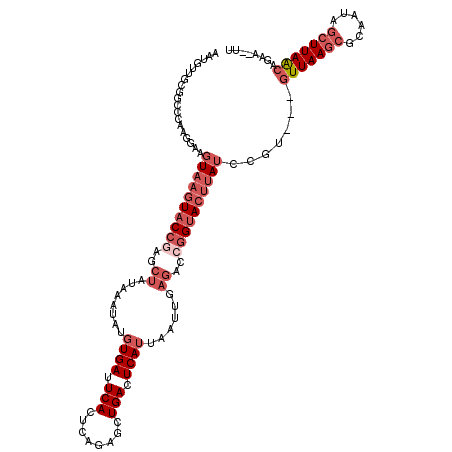
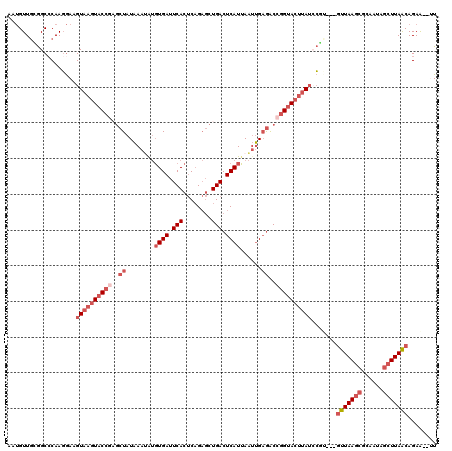
| Location | 3,586,977 – 3,587,092 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -17.25 |
| Energy contribution | -19.28 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3586977 115 - 27905053 AA--UUCUGUUAAGCUAUUGCGCUUAAC---ACGGAUAAGUACCGGUCUCAAUUAAUGAGUCAGCUCUGAGUGAAUCACAUAUUUAUAGCUCGGUACUUACUUCCUUGUGCCGCAACAUU ..--...((((((((......)))))))---).(((((((((((((.((((.....)))).)((((.((((((.......)))))).)))))))))))))..))).(((......))).. ( -34.20) >DroSec_CAF1 37510 115 - 1 AA--UUCUGCUAAGCUAUUGCGCUUAAC---ACGGAUAAGUACCGGUCUCAAUUAAUGAGUCAGCUCUGAGUGAAUCACAUAUUUAUAGCUCGGUACUUACUUCCUUGGGCCGCAACAUU ..--...(((...((....))((((((.---..(((((((((((((.((((.....)))).)((((.((((((.......)))))).)))))))))))))..))))))))).)))..... ( -32.90) >DroSim_CAF1 38312 115 - 1 AA--UUCUGCUAAGCUAUUGCGCUUAAC---ACGGAUAAGUACCGGUCUCAAUUAAUGAGUCAGCUCUGAGUGAAUCACAUAUUUAUAGCGCGGUACUUACUUCCUUGGGCCGCAACAUU ..--...(((...((....))((((((.---..(((((((((((((.((((.....)))).).(((.((((((.......)))))).))).)))))))))..))))))))).)))..... ( -32.90) >DroEre_CAF1 43111 115 - 1 AU--UUCUGUUAAGCUAUUGCGCUUAAC---CAGUAUAAGUACCGGUCUCAAUUAAUGAGUCAGCUCUGAGUGAAUCACACGUUUAUAGCUCGGUACUUACUUCCUUGGGCCCCAACAUU ..--..(((((((((......)))))).---)))..((((((((((.((((.....)))).)((((.((((((.....)))..))).))))))))))))).....((((...)))).... ( -29.70) >DroYak_CAF1 60751 115 - 1 AU--UUCUGUUAAGCUGUUGCGCUUAAC---GAGUCUAAGUACCGGUCUCAAUUAAUGAGUCAGCUCGGAGUGAAUCACAUAUUUAUAGCUGGGUACUUACUUCCUUGGGCCUCAACAUU ..--...........(((((.((((((.---(((..((((((((...((((.....)))).(((((..(((((.......)))))..))))))))))))).))).))))))..))))).. ( -31.00) >DroAna_CAF1 34402 111 - 1 AACUCUCUGUUAA-----UUCCCUUAAUGACGUCGAUAGCUGCCGGCUACUGAUCAUGAGUCAGUGCUGGGUGAUUCAGGUUUUAAGA----UGUACUAACUUUGUUGGGCUUGAACUGU .............-----...(((.(((.((((.....))..(((((.((((((.....))))))))))))).))).))).((((((.----....(((((...))))).)))))).... ( -24.80) >consensus AA__UUCUGUUAAGCUAUUGCGCUUAAC___ACGGAUAAGUACCGGUCUCAAUUAAUGAGUCAGCUCUGAGUGAAUCACAUAUUUAUAGCUCGGUACUUACUUCCUUGGGCCGCAACAUU ........(((((((......))))))).....(((((((((((((.((((.....)))).).(((.((((((.......)))))).))).)))))))))..)))............... (-17.25 = -19.28 + 2.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:17 2006