
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,583,275 – 3,583,429 |
| Length | 154 |
| Max. P | 0.999913 |

| Location | 3,583,275 – 3,583,389 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3583275 114 + 27905053 UCUUCUGCUUC-CAUUUACAUCACUACUUAUGGCAAGCUGCAUCGCAUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAA ......(((((-(((..............)))).)))).......((((((((....((((((((((.((..(((.....)))..)).))))))))))....))))))))..... ( -32.54) >DroSec_CAF1 34125 114 + 1 UCUUCGGCUUC-CAUUUACAUCACUACUUAUGGCAAGCUGCAUCGCAUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCACUGGGGAUGAACAUACAAAUUUCAUGGUAAA ....(((((((-(((..............)))).)))))).....((((((((....((((((((((.((..(((.....)))..)).))))))))))....))))))))..... ( -36.04) >DroSim_CAF1 34820 114 + 1 UCUCCUCCUUC-CAUUUAAAUCACUACUUAUGGCAAGCUGCAUCGCUUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAA ..........(-(((.............(((..(((((......)))))..)))...((((((((((.((..(((.....)))..)).)))))))))).........)))).... ( -26.50) >DroEre_CAF1 39478 114 + 1 UCUUCUUUUCU-UUUUUACAUCACUACUUAUGGCAAGCUGCAUCUCAUGAAAUGCAAAUGUUUAUCUGCAACGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAA ...........-..(((((.............((.....))....((((((((....((((((((((.((.((((.....)))).)).))))))))))....))))))))))))) ( -32.10) >DroYak_CAF1 26538 115 + 1 UCUUCUUUUCUCUUUUUACAUCACUACUUAUGGCAAGCUGCAUCUCAUGAAAUGCAAAUGUAUAUCUGUAAGGACUCAAAGUCGCUGUGGAUGAACAUACAAAUUUUAUGGUAAA ..............(((((.............((.....))....((((((((....((((.((((..((..(((.....)))..))..)))).))))....))))))))))))) ( -22.00) >consensus UCUUCUGCUUC_CAUUUACAUCACUACUUAUGGCAAGCUGCAUCGCAUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAA ..............(((((.............((.....))....((((((((....((((((((((.((..(((.....)))..)).))))))))))....))))))))))))) (-25.94 = -26.22 + 0.28)

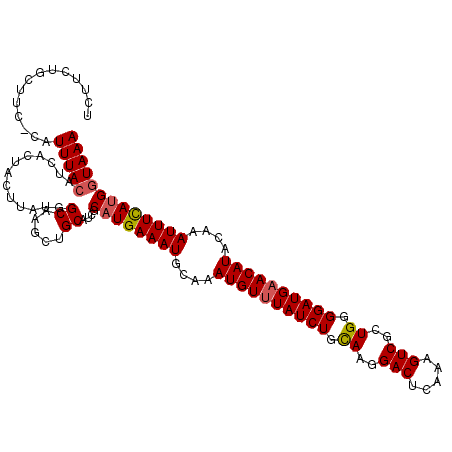
| Location | 3,583,309 – 3,583,429 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -33.19 |
| Consensus MFE | -30.26 |
| Energy contribution | -29.50 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3583309 120 + 27905053 AGCUGCAUCGCAUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAACUCUCUGAUCGCUCCUGCAAUUGGGAUAAAAGGAAGCAAG ...(((.((.((((((((....((((((((((.((..(((.....)))..)).))))))))))....))))))))......((.(..((.((....)).))..)))......)).))).. ( -35.60) >DroSec_CAF1 34159 120 + 1 AGCUGCAUCGCAUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCACUGGGGAUGAACAUACAAAUUUCAUGGUAAACUCUUUGAUCGCUCCAACAAUUUGGAUAAAAGGAAGCAAG .(((......((((((((....((((((((((.((..(((.....)))..)).))))))))))....))))))))......(((((......(((((....)))))...))))))))... ( -35.40) >DroSim_CAF1 34854 120 + 1 AGCUGCAUCGCUUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAACUCUUUGAUCGCUCCAACAAUUUGGAUAAAAGGAAGCAAG .(((.....(((((((((....((((((((((.((..(((.....)))..)).))))))))))....)))))).)))....(((((......(((((....)))))...))))))))... ( -31.90) >DroEre_CAF1 39512 120 + 1 AGCUGCAUCUCAUGAAAUGCAAAUGUUUAUCUGCAACGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAACUCUCUGAUCUCUCCAGCAAUUUGGAUAAAAGGAAGCGAA .(((.....(((((((((....((((((((((.((.((((.....)))).)).))))))))))....))))))))).......(((......(((((....))))).....))))))... ( -36.80) >DroYak_CAF1 26573 119 + 1 AGCUGCAUCUCAUGAAAUGCAAAUGUAUAUCUGUAAGGACUCAAAGUCGCUGUGGAUGAACAUACAAAUUUUAUGGUAAACUCUCUGAUCGCUUCAAUAAUUUAGAUAAA-GGGAGCAAA ...(((...(((((((((....((((.((((..((..(((.....)))..))..)))).))))....))))))))).....(((((.(((..............)))..)-))))))).. ( -26.24) >consensus AGCUGCAUCGCAUGAAAUGCAAAUGUUUAUCUGCAAGGACUCAAAGUCGCUGGGGAUGAACAUACAAAUUUCAUGGUAAACUCUCUGAUCGCUCCAACAAUUUGGAUAAAAGGAAGCAAG ...(((.((.((((((((....((((((((((.((..(((.....)))..)).))))))))))....)))))))).................(((((....)))))......)).))).. (-30.26 = -29.50 + -0.76)

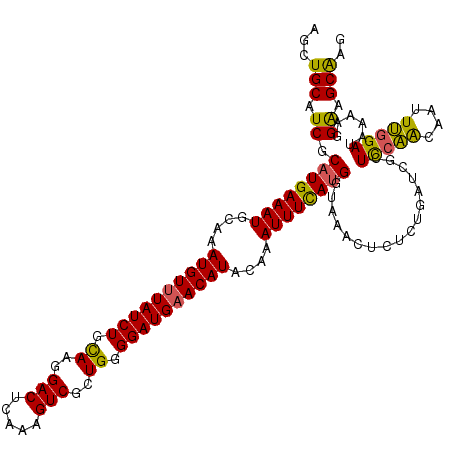

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:14 2006