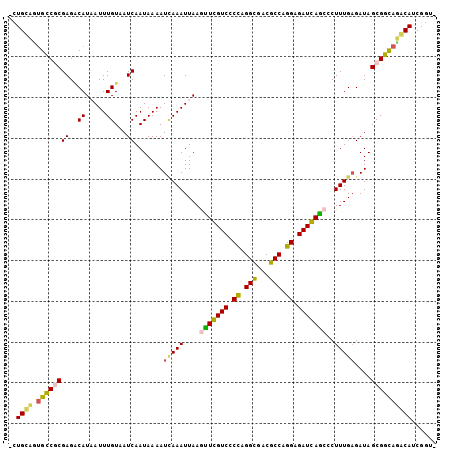
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 672,388 – 672,506 |
| Length | 118 |
| Max. P | 0.875216 |

| Location | 672,388 – 672,492 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
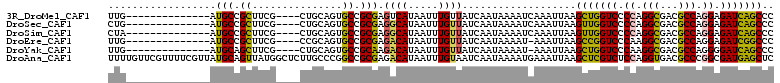
>3R_DroMel_CAF1 672388 104 + 27905053 GACCGAUGCUUGCCGCUAUCUCAAAGGGCUGAUCUCCUGGCGUCGCCUGGGGACCAGCUUAAUUUGAUUUUAUUGAUAACAAAUUAUGACUCGCGGCACUGCAG- ......(((.((((((....(((((.(((((.(((((.(((...))).))))).)))))...))))).....(((....)))..........))))))..))).- ( -36.60) >DroPse_CAF1 93021 104 + 1 -UCCGUUGUCUACCGCUAUCUCAAAGGGUUCAUCUCCAGGCGUCGCCUAGAGACGAAUUUAAUUUAAUUUUAUUGAUUACAAAUGAUGUCUCGAGACAAGACAAC -...(((((((.........(((..((((((.((((.((((...)))).)))).))))))....(((((.....)))))....)))((((....))))))))))) ( -26.20) >DroSec_CAF1 91753 104 + 1 GACCGAUGCAUGCCGCUAUCUCAAAGGGCUGAUCUCCUGGCGUCGCCUGGGGACCAACUUAAUUUGAUUUUAUUGAUAACAAAUUAUGCCUCGCGGCACUGCAG- ......((((((((((.........((((((.(((((.(((...))).))))).))...(((((((.((........))))))))).)))).)))))).)))).- ( -32.60) >DroSim_CAF1 89464 104 + 1 GACCGAUGCAUGCCGCUAUCUCAAAGGGCUGAUCUCCUGGCGUCGCCUGGGGACCAACUUAAUUUGAUUUUAUUGAUAACAAAUUAUGCCUCGCGGCACUGCAG- ......((((((((((.........((((((.(((((.(((...))).))))).))...(((((((.((........))))))))).)))).)))))).)))).- ( -32.60) >DroAna_CAF1 91858 103 + 1 --UCGAUGUCUGCCGCUAUCACAAAGAGCUCAUCGCCGGGCGUCACCUGGAGACGAGCUUAAUUUCAUUUUAUUGAUUACAAAUUAUGUCUCGCGGCCGGGCAAG --....((((((((((....(((..((((((.((.(((((.....))))).)).))))))....(((......)))..........)))...)))).)))))).. ( -35.10) >DroPer_CAF1 94064 104 + 1 -UCCGUUGUCUACCGCUAUCUCAAAGGGUUCAUCUCCAGGCGUCGCCUAGAGACGAAUUUAAUUUAAUUUUAUUGAUUACAAAUGAUGUCUCGAGACAAGACAAC -...(((((((.........(((..((((((.((((.((((...)))).)))).))))))....(((((.....)))))....)))((((....))))))))))) ( -26.20) >consensus _ACCGAUGCCUGCCGCUAUCUCAAAGGGCUCAUCUCCUGGCGUCGCCUGGAGACCAACUUAAUUUGAUUUUAUUGAUAACAAAUUAUGUCUCGCGGCACGGCAA_ ......((((((((((.........((((((.(((((.(((...))).))))).))))))............(((....)))..........)))))).)))).. (-23.70 = -23.98 + 0.28)

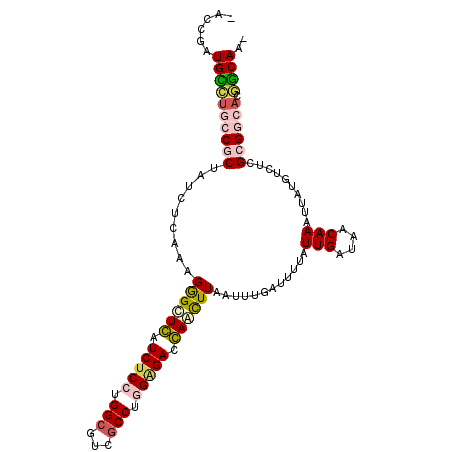
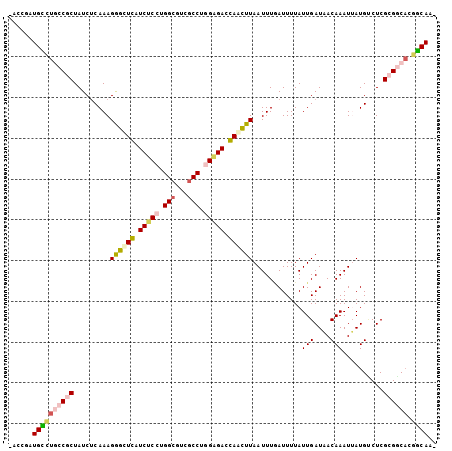
| Location | 672,388 – 672,492 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -26.30 |
| Energy contribution | -25.22 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
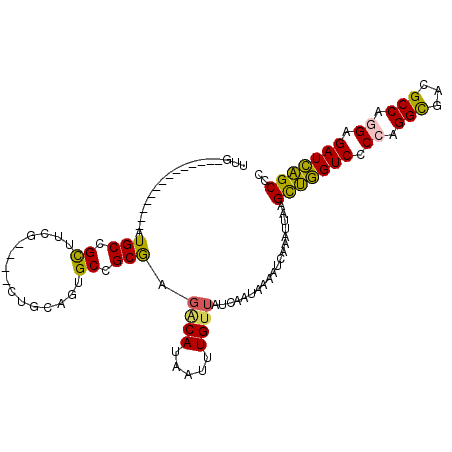
>3R_DroMel_CAF1 672388 104 - 27905053 -CUGCAGUGCCGCGAGUCAUAAUUUGUUAUCAAUAAAAUCAAAUUAAGCUGGUCCCCAGGCGACGCCAGGAGAUCAGCCCUUUGAGAUAGCGGCAAGCAUCGGUC -.(((..((((((..(((.(((((((.............))))))).(((((((.((.(((...))).)).))))))).......))).)))))).)))...... ( -36.52) >DroPse_CAF1 93021 104 - 1 GUUGUCUUGUCUCGAGACAUCAUUUGUAAUCAAUAAAAUUAAAUUAAAUUCGUCUCUAGGCGACGCCUGGAGAUGAACCCUUUGAGAUAGCGGUAGACAACGGA- (((((((((((((((((((.....))).....................(((((((((((((...)))))))))))))...))))))))).....)))))))...- ( -36.90) >DroSec_CAF1 91753 104 - 1 -CUGCAGUGCCGCGAGGCAUAAUUUGUUAUCAAUAAAAUCAAAUUAAGUUGGUCCCCAGGCGACGCCAGGAGAUCAGCCCUUUGAGAUAGCGGCAUGCAUCGGUC -.((((.((((((((((((.....)))).)).......(((((....(((((((.((.(((...))).)).)))))))..)))))....))))))))))...... ( -35.40) >DroSim_CAF1 89464 104 - 1 -CUGCAGUGCCGCGAGGCAUAAUUUGUUAUCAAUAAAAUCAAAUUAAGUUGGUCCCCAGGCGACGCCAGGAGAUCAGCCCUUUGAGAUAGCGGCAUGCAUCGGUC -.((((.((((((((((((.....)))).)).......(((((....(((((((.((.(((...))).)).)))))))..)))))....))))))))))...... ( -35.40) >DroAna_CAF1 91858 103 - 1 CUUGCCCGGCCGCGAGACAUAAUUUGUAAUCAAUAAAAUGAAAUUAAGCUCGUCUCCAGGUGACGCCCGGCGAUGAGCUCUUUGUGAUAGCGGCAGACAUCGA-- ........(((((....(((((.......(((......))).....((((((((.((.((.....)).)).))))))))..)))))...))))).........-- ( -31.60) >DroPer_CAF1 94064 104 - 1 GUUGUCUUGUCUCGAGACAUCAUUUGUAAUCAAUAAAAUUAAAUUAAAUUCGUCUCUAGGCGACGCCUGGAGAUGAACCCUUUGAGAUAGCGGUAGACAACGGA- (((((((((((((((((((.....))).....................(((((((((((((...)))))))))))))...))))))))).....)))))))...- ( -36.90) >consensus _CUGCAGUGCCGCGAGACAUAAUUUGUAAUCAAUAAAAUCAAAUUAAGUUCGUCCCCAGGCGACGCCAGGAGAUCAGCCCUUUGAGAUAGCGGCAGACAUCGGU_ ..((((.((((((((..((.....))...)).......(((((....(((((((.((.(((...))).)).)))))))..)))))....))))))))))...... (-26.30 = -25.22 + -1.08)

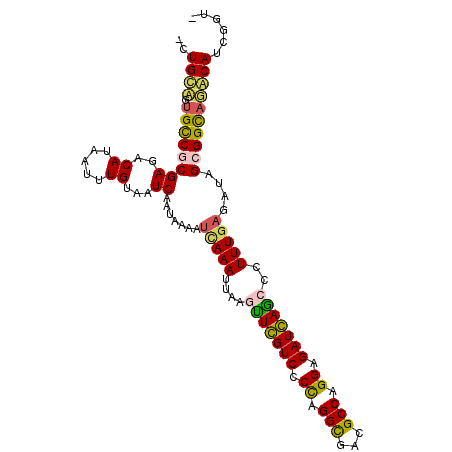
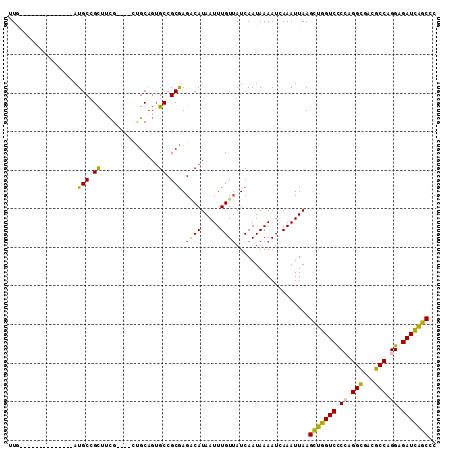
| Location | 672,413 – 672,506 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -19.52 |
| Energy contribution | -18.83 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 672413 93 - 27905053 UUG--------------AUGCCGCUUCG----CUGCAGUGCCGCGAGUCAUAAUUUGUUAUCAAUAAAAUCAAAUUAAGCUGGUCCCCAGGCGACGCCAGGAGAUCAGCCC .((--------------((..(((..((----(....)))..))).))))(((((((.............))))))).(((((((.((.(((...))).)).))))))).. ( -28.72) >DroSec_CAF1 91778 93 - 1 CUG--------------AUGCCGCUUCG----CUGCAGUGCCGCGAGGCAUAAUUUGUUAUCAAUAAAAUCAAAUUAAGUUGGUCCCCAGGCGACGCCAGGAGAUCAGCCC ...--------------.....((((((----(.((...)).))))))).(((((((.............))))))).(((((((.((.(((...))).)).))))))).. ( -30.52) >DroSim_CAF1 89489 93 - 1 CUA--------------AUGCCGCUUCG----CUGCAGUGCCGCGAGGCAUAAUUUGUUAUCAAUAAAAUCAAAUUAAGUUGGUCCCCAGGCGACGCCAGGAGAUCAGCCC ...--------------.....((((((----(.((...)).))))))).(((((((.............))))))).(((((((.((.(((...))).)).))))))).. ( -30.52) >DroEre_CAF1 88422 92 - 1 UUG--------------AUGCCGCUUCG----CCGCAGUGCCGCGAGACAUAAUUUGUUAUCAAUAAAAU-AAAUUAAGCCGGUCCCAAGGCGACGCCAGGAGAUCGGCCC ...--------------.....(..(((----(.((...)).))))..).(((((((((........)))-)))))).(((((((((..(((...))).)).))))))).. ( -27.80) >DroYak_CAF1 100557 92 - 1 UUG--------------AUGCAGCUUCG----CUGCAGUGCCGCAAGACAUAAUUUGUUAUCAAUAAAAU-AAAUUAAGCUGGUCCCAAGGCGACGCCAGGGGAUCAGCCC ...--------------.((((((...)----)))))(((.(....).)))((((((((........)))-)))))..(((((((((..(((...)))..))))))))).. ( -34.80) >DroAna_CAF1 91881 111 - 1 UUUUGUUCGUUUUCGUUAUGCAGUUAUGGCUCUUGCCCGGCCGCGAGACAUAAUUUGUAAUCAAUAAAAUGAAAUUAAGCUCGUCUCCAGGUGACGCCCGGCGAUGAGCUC .....((((((((.((((((.(((((((..((((((......)))))))))))))))))))....))))))))....((((((((.((.((.....)).)).)))))))). ( -34.90) >consensus UUG______________AUGCCGCUUCG____CUGCAGUGCCGCGAGACAUAAUUUGUUAUCAAUAAAAUCAAAUUAAGCUGGUCCCCAGGCGACGCCAGGAGAUCAGCCC ..................(((.((...............)).))).((((.....))))...................(((((((.((.(((...))).)).))))))).. (-19.52 = -18.83 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:40 2006