| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
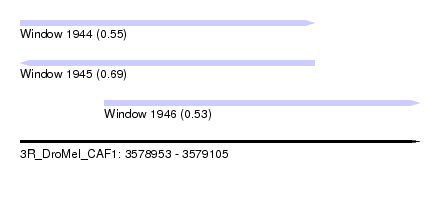
| Location | 3,578,953 – 3,579,105 |
| Length | 152 |
| Max. P | 0.691084 |

| Location | 3,578,953 – 3,579,065 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -13.94 |
| Energy contribution | -13.56 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
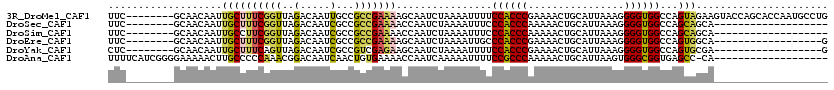
>3R_DroMel_CAF1 3578953 112 + 27905053 UUC--------GCAACAAUUGCUUUCGGUUAGACAAUUGCCGCCGAAAAGCAAUCUAAAAUUUUCCACCCGAAAACUGCAUUAAAGGGGUGGCCAGUAGAAGUACCAGCACCAAUGCCUG ...--------((.....(((((((((((............)))).)))))))((((.......((((((................))))))....)))).......))........... ( -25.09) >DroSec_CAF1 29888 93 + 1 UUC--------GCAACAAUUGCUUUCGGUUAGACAAUCGCCGCCGAAAACCAAUCUAAAAUUUCCCACCCAAAAACUGCAUUAAAGGGGUGGCCAGCAGCA------------------- ...--------((....((((.(((((((..(.....)...)))))))..))))..........((((((................))))))...))....------------------- ( -19.99) >DroSim_CAF1 30370 93 + 1 UUC--------GCAACAAUUGCCUUCGGUUAGACAAUCGCCGCCGAAAACCAAUCUAAAAUUUCCCACCCAAAAACUGCAUUAAAGGGGUGGCCAGCAGCA------------------- ...--------((....((((..((((((..(.....)...))))))...))))..........((((((................))))))...))....------------------- ( -19.09) >DroEre_CAF1 35244 94 + 1 UUC--------GCAACAAUUGCUUUCGGUUAGACAAUCGCCGCCGAAAAGCAAUCUAAAAUUGCCCACCCGAAAACUGCAUUAAAGGGGUGGCCAGUGGCA------------------G ...--------((....((((((((((((..(.....)...)))).)))))))).....((((.((((((................)))))).)))).)).------------------. ( -28.19) >DroYak_CAF1 22123 94 + 1 CUC--------GCAACAAUUGCUUUCAGUUAGACAAUCGCCGUCGAGAAGCAAUCUAAAAUUUUCCACCCGAAAACUGCAUUAAAGGGGUGGCCAGUGCGA------------------G (((--------(((...((((((((......(((.......)))..))))))))..........((((((................))))))....)))))------------------) ( -25.59) >DroAna_CAF1 25705 100 + 1 UUUUCAUCGGGGAAAAACUUGCCCCCAAACGGACAAUCAACUGUGAAAACCAAUCAAAAAUUUUCCGCCCAAAAACUGCAUUAAGUGGGCGGUGAGCC-CA------------------- (((((...((((..........))))..((((........)))))))))...........(((.((((((....(((......))))))))).)))..-..------------------- ( -23.80) >consensus UUC________GCAACAAUUGCUUUCGGUUAGACAAUCGCCGCCGAAAACCAAUCUAAAAUUUCCCACCCAAAAACUGCAUUAAAGGGGUGGCCAGCAGCA___________________ ...................((((((((((..(.....)...)))))))................((((((................))))))...)))...................... (-13.94 = -13.56 + -0.38)
| Location | 3,578,953 – 3,579,065 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -18.99 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
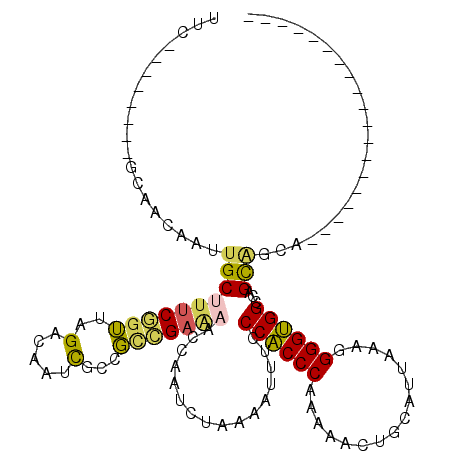
>3R_DroMel_CAF1 3578953 112 - 27905053 CAGGCAUUGGUGCUGGUACUUCUACUGGCCACCCCUUUAAUGCAGUUUUCGGGUGGAAAAUUUUAGAUUGCUUUUCGGCGGCAAUUGUCUAACCGAAAGCAAUUGUUGC--------GAA ...((((((((((..(((....)))..).)))).....))))).((((((((.((((........(((((((.......))))))).)))).)))))))).........--------... ( -34.00) >DroSec_CAF1 29888 93 - 1 -------------------UGCUGCUGGCCACCCCUUUAAUGCAGUUUUUGGGUGGGAAAUUUUAGAUUGGUUUUCGGCGGCGAUUGUCUAACCGAAAGCAAUUGUUGC--------GAA -------------------.((.((...((((((................)))))).........(((((.(((((((.(((....)))...)))))))))))))).))--------... ( -24.49) >DroSim_CAF1 30370 93 - 1 -------------------UGCUGCUGGCCACCCCUUUAAUGCAGUUUUUGGGUGGGAAAUUUUAGAUUGGUUUUCGGCGGCGAUUGUCUAACCGAAGGCAAUUGUUGC--------GAA -------------------.(((((((.((((((................))))))((((((.......)))))))))))))((((((((......)))))))).....--------... ( -28.79) >DroEre_CAF1 35244 94 - 1 C------------------UGCCACUGGCCACCCCUUUAAUGCAGUUUUCGGGUGGGCAAUUUUAGAUUGCUUUUCGGCGGCGAUUGUCUAACCGAAAGCAAUUGUUGC--------GAA .------------------(((.((((.((((((................)))))).))......(((((((((.(((.(((....)))...)))))))))))))).))--------).. ( -29.89) >DroYak_CAF1 22123 94 - 1 C------------------UCGCACUGGCCACCCCUUUAAUGCAGUUUUCGGGUGGAAAAUUUUAGAUUGCUUCUCGACGGCGAUUGUCUAACUGAAAGCAAUUGUUGC--------GAG (------------------(((((....((((((................)))))).........((((((((.(((..(((....)))....)))))))))))..)))--------))) ( -26.39) >DroAna_CAF1 25705 100 - 1 -------------------UG-GGCUCACCGCCCACUUAAUGCAGUUUUUGGGCGGAAAAUUUUUGAUUGGUUUUCACAGUUGAUUGUCCGUUUGGGGGCAAGUUUUUCCCCGAUGAAAA -------------------.(-(((...(((((((..............))))))).......(..((((.......))))..)..))))..(((((((........)))))))...... ( -31.14) >consensus ___________________UGCUACUGGCCACCCCUUUAAUGCAGUUUUCGGGUGGAAAAUUUUAGAUUGCUUUUCGGCGGCGAUUGUCUAACCGAAAGCAAUUGUUGC________GAA ............................((((((................)))))).........(((((.(((((((.(((....)))...))))))))))))................ (-18.99 = -18.57 + -0.41)


| Location | 3,578,985 – 3,579,105 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.56 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3578985 120 + 27905053 CGCCGAAAAGCAAUCUAAAAUUUUCCACCCGAAAACUGCAUUAAAGGGGUGGCCAGUAGAAGUACCAGCACCAAUGCCUGUGCCGUAAAUUAAUAUGGUGGCGAAACUCUAAAAUUGAAA ((((.....(((.((((.......((((((................))))))....)))).......(((....))).)))((((((......))))))))))................. ( -25.89) >DroSec_CAF1 29920 99 + 1 CGCCGAAAACCAAUCUAAAAUUUCCCACCCAAAAACUGCAUUAAAGGGGUGGCCAGCAGCA---------------------CCGUAAAUUAAUAUGGUGGCGAAAGUCUAAAAUUGAAA ((((....................((((((................))))))........(---------------------(((((......))))))))))................. ( -21.59) >DroSim_CAF1 30402 99 + 1 CGCCGAAAACCAAUCUAAAAUUUCCCACCCAAAAACUGCAUUAAAGGGGUGGCCAGCAGCA---------------------CCGUAAAUUAAUAUGGUGGCGAAAGUCUAAAAUUGAAA ((((....................((((((................))))))........(---------------------(((((......))))))))))................. ( -21.59) >DroEre_CAF1 35276 102 + 1 CGCCGAAAAGCAAUCUAAAAUUGCCCACCCGAAAACUGCAUUAAAGGGGUGGCCAGUGGCA------------------GUGUCGUAAAUUAAUAUGGUGGCGAAACUCUAAAAUUGAAA .((((....(((((.....)))))((((((................))))))....))))(------------------((.((((...(......)...)))).)))............ ( -22.59) >DroYak_CAF1 22155 102 + 1 CGUCGAGAAGCAAUCUAAAAUUUUCCACCCGAAAACUGCAUUAAAGGGGUGGCCAGUGCGA------------------GUGCCGUAAAUUAAUAUGGUGGCGAAACUGUAAAAUUGAAA ..((((..................((((((................)))))).((((.((.------------------.(((((((......))))))).))..)))).....)))).. ( -20.99) >DroAna_CAF1 25745 98 + 1 CUGUGAAAACCAAUCAAAAAUUUUCCGCCCAAAAACUGCAUUAAGUGGGCGGUGAGCC-CA---------------------CCAUAAAUUAAUGUGCUGGCAAAACUAUAAAAUUGAAA ..((....))...((((.......((((((....(((......)))))))))...(((-..---------------------.((((......))))..)))............)))).. ( -20.60) >consensus CGCCGAAAACCAAUCUAAAAUUUCCCACCCAAAAACUGCAUUAAAGGGGUGGCCAGCAGCA_____________________CCGUAAAUUAAUAUGGUGGCGAAACUCUAAAAUUGAAA (((((...................((((((................))))))..............................(((((......))))))))))................. (-16.80 = -16.56 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:11 2006