
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,550,751 – 3,550,861 |
| Length | 110 |
| Max. P | 0.747097 |

| Location | 3,550,751 – 3,550,861 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
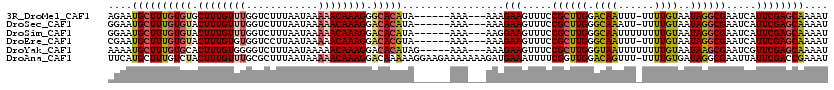
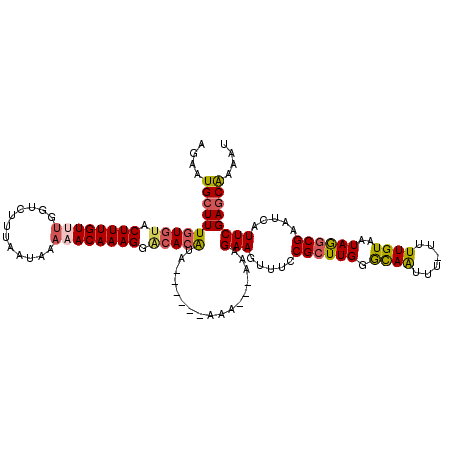
>3R_DroMel_CAF1 3550751 110 - 27905053 AGAAUGCUUUGUGUGCUUUGUUUGGUCUUUAAUAAAAACAAAGGGCACAUA------AAA---AAAGAAGUUUCCGCUUGGACAAUUU-UUUUGUAAUAGGCGAAUCAUUCGAGCAAAAU ....((((((((((.((((((((............)))))))).)))))..------...---...(((.....((((((.((((...-..))))..)))))).....)))))))).... ( -29.80) >DroSec_CAF1 9624 110 - 1 GGAAUGCUUUGUGUACUUUGUUUGGUCUUUAAUAAAAACAAAGGACACAUA------AAA---AAAGAAGUUUCCGCUUGGGCAAAUU-UUUUGUAAUAGGCGAAUCAUUCGAGCAAAAU ....((((((((((.((((((((............)))))))).)))))..------...---...(((.....((((((.((((...-..))))..)))))).....)))))))).... ( -26.90) >DroSim_CAF1 9888 111 - 1 GGAAUGCUUUGUGUACUUUGUUUGGUCUUUAAUAAAAACAAAGGACACAUA------AAA---AAGGAAGUUUCCGCUUGGGCAAUUUUUUUUGUAAUAGGCGAAUCAUUCGAGCAAAAU ....((((((((((.((((((((............)))))))).)))))..------...---...(((.....((((((.((((......))))..)))))).....)))))))).... ( -27.70) >DroEre_CAF1 10069 110 - 1 CGAAUGCUUUGUGUACUUUGUGUGGUCCUUAAUAAAAACAAAGGACACGUA------AAA---AAAGAAGUUUCCGCUUGGGCAAUUU-UUUUGUAAUAGGCGAAUCAUUCGAGCAAAAU ((((((.(((((.((.((((((((.(((((..........)))))))))))------)).---(((((((((.((.....)).)))))-))))....)).))))).))))))........ ( -27.30) >DroYak_CAF1 10454 112 - 1 AAAAUGCUUUGUGCACUUUGUGGGGUCUUUAAUAAAAACAAAGGACACAUAG-----AAA---AAAGAAGUUUCCGCUUGGGUAAUUUUUUUUGUAAUAAGCGAAUCGUUCGAGCAAAAU ......((.((((..((((((................))))))..)))).))-----...---............(((((((..((((.(((......))).))))..)))))))..... ( -22.49) >DroAna_CAF1 5937 119 - 1 UUCAUGCUUUGUCUACUUUGUUUGCGCUUUAAUAAAAACAAAGGACAAAAAGGAAGAAAAAAAGAUGAAAUUUUCGGUUGGACAGUUU-UUUUGUGAUAGGCGAAUUAUUCGACCGAAAU (((((.((((.(((.((((.(((((.((((.........))))).)))))))).)))...)))))))))..(((((((((((.(((((-.((((...)))).))))).))))))))))). ( -26.90) >consensus AGAAUGCUUUGUGUACUUUGUUUGGUCUUUAAUAAAAACAAAGGACACAUA______AAA___AAAGAAGUUUCCGCUUGGGCAAUUU_UUUUGUAAUAGGCGAAUCAUUCGAGCAAAAU ....((((((((((.((((((((............)))))))).))))).................(((.....((((((.((((......))))..)))))).....)))))))).... (-21.99 = -22.30 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:45 2006