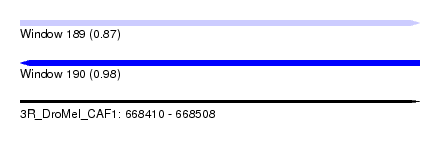
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 668,410 – 668,508 |
| Length | 98 |
| Max. P | 0.983769 |

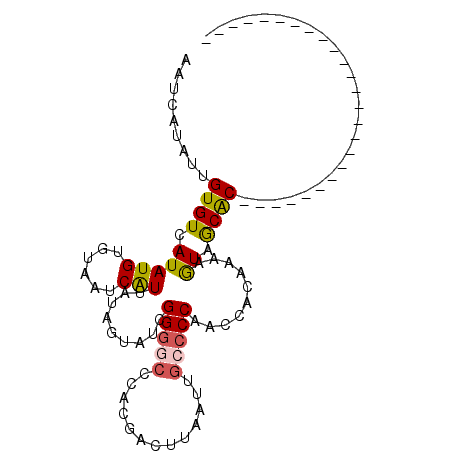
| Location | 668,410 – 668,508 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 71.49 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -8.70 |
| Energy contribution | -8.62 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 668410 98 + 27905053 AAUCAUAUUGUGUCAUAUGUGUAAUCGUUAAAGUAUCGGGGCCCACGACUUAAUUGCCCCGGCCACAAAAGUAACACAUUUGUGUGUUUGU----AUAUGUG .............(((((((...............(((((((.............)))))))..(((((....((((....)))).)))))----))))))) ( -22.22) >DroPse_CAF1 89574 75 + 1 AAUCAUAUUGUGUCAUAUGUCUAAUCAUUAUAUUAUCGGGGCAUACAAUAUAAUUGCCCCAACCACAA--GUGGCAC------------------------- .........(((((((.(((.((((......))))..((((((...........))))))....))).--)))))))------------------------- ( -19.70) >DroSec_CAF1 87846 102 + 1 AAUCAUAUUGUGUCAUAUGUGUAAUCGUUAAAGUAUCGGGGCCCACGACUUAAUUGCCCCGACCACAAAAGUAACACAUUUGUAUGUUUGUACAUAUAUGUG .............((((((((((...((((..((.(((((((.............)))))))..)).....))))((((....))))...)))))))))).. ( -24.92) >DroYak_CAF1 96677 85 + 1 AAUCAUAUUGUGUCAUAUGUGUAAUCGUUAUAGCAUCGG-CCCCACGAUUUAAUUACCCCGACCACCGGAAUAGCACAUAU----------------AUGUG .............(((((((((((((((....((....)-)...))))))........(((.....)))......))))))----------------))).. ( -16.50) >DroAna_CAF1 88573 77 + 1 AAUCAUAUUGUGUCAUAUGUGUAAUCAUUGUGUUAUCGGGUCGUGUCACUUAAUUGCCCCAACCACAAAAGUGGCGC------------------------- .........(((((((.((......))(((((.....((((.((........)).))))....)))))..)))))))------------------------- ( -15.50) >DroPer_CAF1 90627 75 + 1 AAUCAUAUUGUGUCAUAUGUCUAAUCAUUAUAUUAUCGGGGCAUACAAUAUAAUUGCCCCAACCACAA--GUGGCAC------------------------- .........(((((((.(((.((((......))))..((((((...........))))))....))).--)))))))------------------------- ( -19.70) >consensus AAUCAUAUUGUGUCAUAUGUGUAAUCAUUAUAGUAUCGGGGCCCACGACUUAAUUGCCCCAACCACAAAAGUAGCAC_________________________ .........((((.(((((......))).........(((((.............)))))..........)).))))......................... ( -8.70 = -8.62 + -0.08)

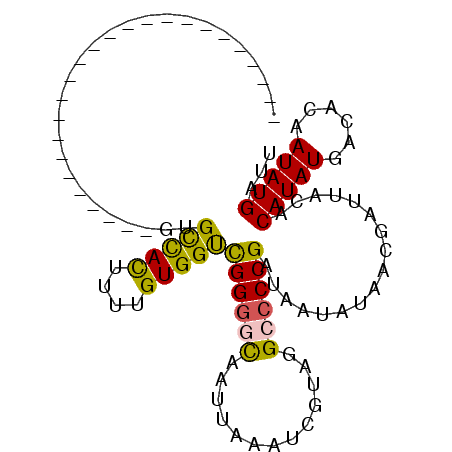
| Location | 668,410 – 668,508 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 71.49 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -14.91 |
| Energy contribution | -14.27 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 668410 98 - 27905053 CACAUAU----ACAAACACACAAAUGUGUUACUUUUGUGGCCGGGGCAAUUAAGUCGUGGGCCCCGAUACUUUAACGAUUACACAUAUGACACAAUAUGAUU ((((...----...((((((....)))))).....))))..((((((.((......))..)))))).................(((((......)))))... ( -23.10) >DroPse_CAF1 89574 75 - 1 -------------------------GUGCCAC--UUGUGGUUGGGGCAAUUAUAUUGUAUGCCCCGAUAAUAUAAUGAUUAGACAUAUGACACAAUAUGAUU -------------------------(((.((.--.(((.(((((((((((......)).)))))))))(((......)))..)))..)).)))......... ( -18.20) >DroSec_CAF1 87846 102 - 1 CACAUAUAUGUACAAACAUACAAAUGUGUUACUUUUGUGGUCGGGGCAAUUAAGUCGUGGGCCCCGAUACUUUAACGAUUACACAUAUGACACAAUAUGAUU ..(((((.((((......))))..(((((((....((((((((((((.((......))..)))))))).(......)....))))..))))))))))))... ( -28.00) >DroYak_CAF1 96677 85 - 1 CACAU----------------AUAUGUGCUAUUCCGGUGGUCGGGGUAAUUAAAUCGUGGGG-CCGAUGCUAUAACGAUUACACAUAUGACACAAUAUGAUU ....(----------------(((((((.(((((((.....)))))))....((((((..((-(....)))...)))))).))))))))............. ( -22.90) >DroAna_CAF1 88573 77 - 1 -------------------------GCGCCACUUUUGUGGUUGGGGCAAUUAAGUGACACGACCCGAUAACACAAUGAUUACACAUAUGACACAAUAUGAUU -------------------------.........(((((((((((.................))))))..)))))........(((((......)))))... ( -13.53) >DroPer_CAF1 90627 75 - 1 -------------------------GUGCCAC--UUGUGGUUGGGGCAAUUAUAUUGUAUGCCCCGAUAAUAUAAUGAUUAGACAUAUGACACAAUAUGAUU -------------------------(((.((.--.(((.(((((((((((......)).)))))))))(((......)))..)))..)).)))......... ( -18.20) >consensus _________________________GUGCCACUUUUGUGGUCGGGGCAAUUAAAUCGUAGGCCCCGAUAAUAUAACGAUUACACAUAUGACACAAUAUGAUU ...........................(((((....)))))((((((.............)))))).................(((((......)))))... (-14.91 = -14.27 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:35 2006