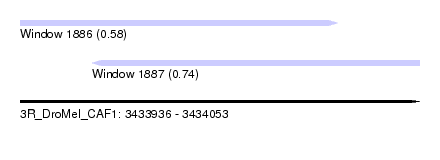
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,433,936 – 3,434,053 |
| Length | 117 |
| Max. P | 0.738003 |

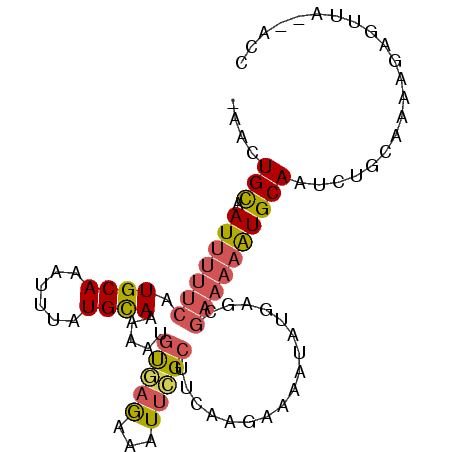
| Location | 3,433,936 – 3,434,029 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -17.83 |
| Consensus MFE | -9.98 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3433936 93 + 27905053 -AACUGCAAUUUUUCAUGCAAAUUUAUGCAAAAAUGUGAGAAAUUCGCUUCAAGAAAAUAUGAGCGAAAAAUGCAAUCUGCAAAAGAGUUA--ACC -(((((((.(((((((((((......))))......)))))))((((((.((........))))))))...)))).(((.....)))))).--... ( -17.90) >DroVir_CAF1 45152 75 + 1 AGCGUGCCAUUUUUCAUGCAGAUUUAUGUAAAAAUUUUUAAAAUUUGCGUAAAGAAAA------------GUGCAAUAGAGA--AAAAU------- ....(((.(((((((((((((((((...((((....))))))))))))))...)))))------------))))).......--.....------- ( -14.40) >DroSim_CAF1 46564 93 + 1 -AACUGCAAUUUUUCAUGCAAAUUUAUGCAAAAAUGUGAGAAAUUCGCUUCAAGAAAAUAUGAGCGAAAAAUGCAAUCUGCAAAAGAGUUA--ACC -(((((((.(((((((((((......))))......)))))))((((((.((........))))))))...)))).(((.....)))))).--... ( -17.90) >DroEre_CAF1 42897 93 + 1 -AACUGCAAUUUUUCAUGCAAAUUUAUGCAAAAAUGUGAGAAAUUCGCUUCAAGAAAAUAUGAGCGAAAAAUGCAAUCUGCAAAAGAGUUA--ACC -(((((((.(((((((((((......))))......)))))))((((((.((........))))))))...)))).(((.....)))))).--... ( -17.90) >DroYak_CAF1 55038 93 + 1 -AACUGCAAUUUUUCAUGCAAAUUUAUGCAAAAAUGUGAGAAAUUCGCUUCAAGAAAAUAUGAGCGAAAAAUGCAAUCUGCAAAAGAGUUA--ACC -(((((((.(((((((((((......))))......)))))))((((((.((........))))))))...)))).(((.....)))))).--... ( -17.90) >DroAna_CAF1 31862 95 + 1 -AACUGUAAUUUUUCAUGCAAAUUUAUGCAGAAAUGCAAGAAAUUUGCUAAAGGAAAAUAUGAGCGAAAAAUGCAAUCUGCAAAAGAGUUAAAACC -((((.(.(((((((..((((((((.((((....))))..))))))))....)))))))............(((.....)))..).))))...... ( -21.00) >consensus _AACUGCAAUUUUUCAUGCAAAUUUAUGCAAAAAUGUGAGAAAUUCGCUUCAAGAAAAUAUGAGCGAAAAAUGCAAUCUGCAAAAGAGUUA__ACC ....(((.(((((((.((((......)))).....(((((...))))).................))))))))))..................... ( -9.98 = -10.40 + 0.42)

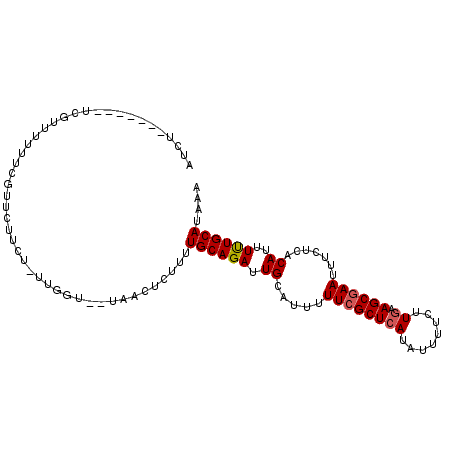
| Location | 3,433,957 – 3,434,053 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 90.87 |
| Mean single sequence MFE | -13.63 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.65 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3433957 96 - 27905053 AUCU-------UCGUUUUUUCGUUCUUCU-UUGGU--UAACUCUUUUGCAGAUUGCAUUUUUCGCUCAUAUUUUCUUGAAGCGAAUUUCUCACAUUUUUGCAUAAA ....-------..(((...(((.......-.))).--.))).....((((((.((.....((((((((........)).)))))).......))..)))))).... ( -13.60) >DroSec_CAF1 32132 96 - 1 AUCU-------UCGUUUUUUCGCUCUUCU-UUGGU--UAACUCUUUUGCAGAUUGCAUUUUUCGCUCAUAUUUUCUUGAAGCGAAUUUCUCACAUUUUUGCAUAAA ....-------..................-.....--.........((((((.((.....((((((((........)).)))))).......))..)))))).... ( -12.90) >DroSim_CAF1 46585 96 - 1 AUCU-------UCGUUUUUUCGCUCUUCU-UUGGU--UAACUCUUUUGCAGAUUGCAUUUUUCGCUCAUAUUUUCUUGAAGCGAAUUUCUCACAUUUUUGCAUAAA ....-------..................-.....--.........((((((.((.....((((((((........)).)))))).......))..)))))).... ( -12.90) >DroEre_CAF1 42918 104 - 1 AUCUGCGUUCUUCUUUUUUUCGUUCUUCUUUUGGU--UAACUCUUUUGCAGAUUGCAUUUUUCGCUCAUAUUUUCUUGAAGCGAAUUUCUCACAUUUUUGCAUAAA (((((((.....)........(((..((....)).--.)))......))))))((((...((((((((........)).)))))).............)))).... ( -15.59) >DroYak_CAF1 55059 96 - 1 GUCU-------UCGUUUUUUCAUUCUUCU-UUGGU--UAACUCUUUUGCAGAUUGCAUUUUUCGCUCAUAUUUUCUUGAAGCGAAUUUCUCACAUUUUUGCAUAAA ....-------..(((...(((.......-.))).--.))).....((((((.((.....((((((((........)).)))))).......))..)))))).... ( -14.30) >DroAna_CAF1 31883 98 - 1 UGCA-------UUGUUUUUUAUUUCUUCU-UUGGUUUUAACUCUUUUGCAGAUUGCAUUUUUCGCUCAUAUUUUCCUUUAGCAAAUUUCUUGCAUUUCUGCAUAAA ....-------..................-................((((((.((((...((.(((.............))).)).....))))..)))))).... ( -12.52) >consensus AUCU_______UCGUUUUUUCGUUCUUCU_UUGGU__UAACUCUUUUGCAGAUUGCAUUUUUCGCUCAUAUUUUCUUGAAGCGAAUUUCUCACAUUUUUGCAUAAA ..............................................((((((.((.....((((((((........)).)))))).......))..)))))).... (-11.45 = -11.65 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:15 2006