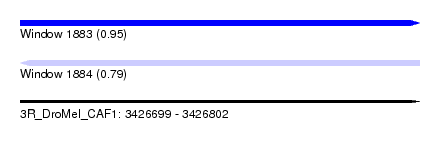
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,426,699 – 3,426,802 |
| Length | 103 |
| Max. P | 0.954137 |

| Location | 3,426,699 – 3,426,802 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -28.94 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
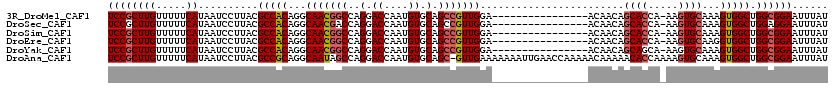
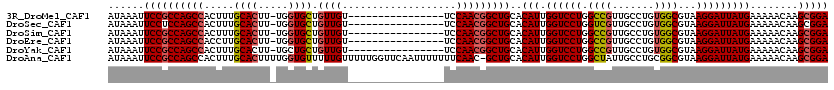
>3R_DroMel_CAF1 3426699 103 + 27905053 UCCGCUUGUUUUUCAUAAUCCUUACGCCACAGGCAACGGCCAGGACCAAUGUGCAGCCGUUGGA----------------ACAACAGCACCA-AAGUGCAAAGUGGCUGGCGGAAUUUAU ((((((((.....))..........(((((...(((((((..(.((....)).).)))))))..----------------......((((..-..))))...))))).))))))...... ( -34.30) >DroSec_CAF1 24895 103 + 1 UCCGCUUGUUUUUCAUAAUCCUUACGCCACAGGCAACGACCAGGACCAAUGUGCAGCCGUUGGA----------------ACAACAGCACCA-AAGUGCAAAGUGGCUGGAGGAAUUUAU ..................(((((..(((((.(....).....(..((((((......)))))).----------------.)....((((..-..))))...)))))..)))))...... ( -28.90) >DroSim_CAF1 39369 103 + 1 UCCGCUUGUUUUUCAUAAUCCUUACGCCACAGGCAACGGCCAGGACCAAUGUGCAGCCGUUGGA----------------ACAACAGCACCA-AAGUGCAAAGUGGCUGGCGGAAUUUAU ((((((((.....))..........(((((...(((((((..(.((....)).).)))))))..----------------......((((..-..))))...))))).))))))...... ( -34.30) >DroEre_CAF1 35551 103 + 1 UCCGCUUGUUUUUCAUAAUCCUUACGCCACAGGCAACGGCCAGGACCAAUGUGCAGCCGUUGGA----------------ACAACAGCACCA-AAGUGCAAGGUGGCUGGCGGAAUUUAU ((((((((.....))..........(((((...(((((((..(.((....)).).)))))))..----------------......((((..-..))))...))))).))))))...... ( -34.10) >DroYak_CAF1 42610 103 + 1 UCCGCUUGUUUUUCAUAAUCCUUACGCCACAGGCAACGGCCAGGACCAAUGUGCAGCCGUUGGA----------------ACAACAGCAGCA-AAGUGCAAAGUGGCUGGCGGAAUUUAU ((((((((.....))..........(((((.(((....)))........((..(.((.((((..----------------....)))).)).-..)..))..))))).))))))...... ( -31.70) >DroAna_CAF1 24106 119 + 1 UCCGCUUGUUUUUCAUAAUCCUUACGCCGCAGGCAAUAGCCAGGACCAAUGUGCAGC-GUUGAAAAAAAUUGAACCAAAAACAAAAACACCAAAAGUGCAAAGUGGCUGGCGGAAUUUAU ((((((((.....))..........(((((.(((....))).((..(((((.....)-)))).......(((.........))).....))...........))))).))))))...... ( -25.60) >consensus UCCGCUUGUUUUUCAUAAUCCUUACGCCACAGGCAACGGCCAGGACCAAUGUGCAGCCGUUGGA________________ACAACAGCACCA_AAGUGCAAAGUGGCUGGCGGAAUUUAU ((((((((.....))..........(((((...(((((((..(.((....)).).)))))))........................((((.....))))...))))).))))))...... (-28.94 = -29.50 + 0.56)
| Location | 3,426,699 – 3,426,802 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -29.41 |
| Energy contribution | -29.83 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.793938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
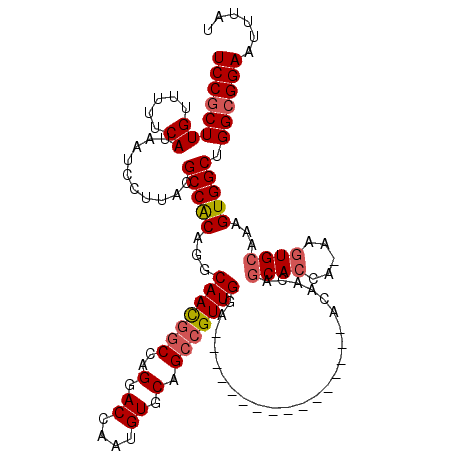
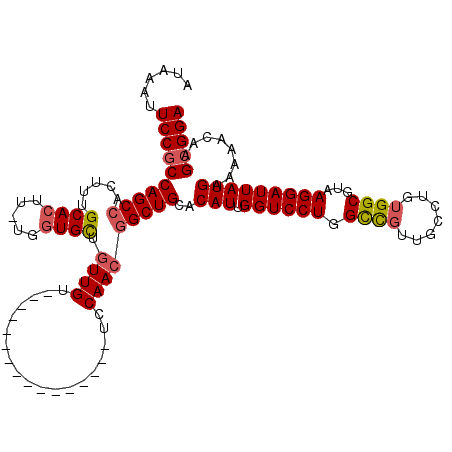
>3R_DroMel_CAF1 3426699 103 - 27905053 AUAAAUUCCGCCAGCCACUUUGCACUU-UGGUGCUGUUGU----------------UCCAACGGCUGCACAUUGGUCCUGGCCGUUGCCUGUGGCGUAAGGAUUAUGAAAAACAAGCGGA ......((((((((((.....((((..-..)))).((((.----------------..)))))))))..(((.((((((.((((.......))))...)))))))))........))))) ( -34.90) >DroSec_CAF1 24895 103 - 1 AUAAAUUCCUCCAGCCACUUUGCACUU-UGGUGCUGUUGU----------------UCCAACGGCUGCACAUUGGUCCUGGUCGUUGCCUGUGGCGUAAGGAUUAUGAAAAACAAGCGGA ......(((..(((((.....((((..-..)))).((((.----------------..)))))))))..(((.((((((...(((((....)))))..)))))))))..........))) ( -29.10) >DroSim_CAF1 39369 103 - 1 AUAAAUUCCGCCAGCCACUUUGCACUU-UGGUGCUGUUGU----------------UCCAACGGCUGCACAUUGGUCCUGGCCGUUGCCUGUGGCGUAAGGAUUAUGAAAAACAAGCGGA ......((((((((((.....((((..-..)))).((((.----------------..)))))))))..(((.((((((.((((.......))))...)))))))))........))))) ( -34.90) >DroEre_CAF1 35551 103 - 1 AUAAAUUCCGCCAGCCACCUUGCACUU-UGGUGCUGUUGU----------------UCCAACGGCUGCACAUUGGUCCUGGCCGUUGCCUGUGGCGUAAGGAUUAUGAAAAACAAGCGGA ......((((((((((.....((((..-..)))).((((.----------------..)))))))))..(((.((((((.((((.......))))...)))))))))........))))) ( -34.90) >DroYak_CAF1 42610 103 - 1 AUAAAUUCCGCCAGCCACUUUGCACUU-UGCUGCUGUUGU----------------UCCAACGGCUGCACAUUGGUCCUGGCCGUUGCCUGUGGCGUAAGGAUUAUGAAAAACAAGCGGA ......(((((..(((((...(((...-(((.(((((((.----------------..))))))).)))....(((....)))..)))..)))))..........((.....)).))))) ( -33.50) >DroAna_CAF1 24106 119 - 1 AUAAAUUCCGCCAGCCACUUUGCACUUUUGGUGUUUUUGUUUUUGGUUCAAUUUUUUUCAAC-GCUGCACAUUGGUCCUGGCUAUUGCCUGCGGCGUAAGGAUUAUGAAAAACAAGCGGA ......(((((..........((((.....))))..((((((((.((..(((((((....((-((((((((.((((....)))).))..))))))))))))))))).))))))))))))) ( -34.70) >consensus AUAAAUUCCGCCAGCCACUUUGCACUU_UGGUGCUGUUGU________________UCCAACGGCUGCACAUUGGUCCUGGCCGUUGCCUGUGGCGUAAGGAUUAUGAAAAACAAGCGGA ......((((((((((.....((((.....)))).((((...................)))))))))..(((.((((((.((((.......))))...)))))))))........))))) (-29.41 = -29.83 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:12 2006